 Your new post is loading...
 Your new post is loading...

|
Scooped by
The Sainsbury Lab
June 5, 2014 6:28 AM
|

|
Scooped by
The Sainsbury Lab
May 7, 2014 8:10 AM
|

|
Rescooped by
The Sainsbury Lab
from Plants and Microbes
April 28, 2014 12:16 PM
|
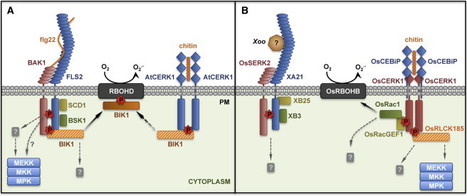
Despite being sessile organisms constantly exposed to potential pathogens and pests, plants are surprisingly resilient to infections. Plants can detect invaders via the recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs). Plant PRRs are surface-localized receptor-like kinases, which comprise a ligand-binding ectodomain and an intracellular kinase domain, or receptor-like proteins, which do not exhibit any known intracellular signaling domain. In this review, we summarize recent discoveries that shed light on the molecular mechanisms underlying ligand perception and subsequent activation of plant PRRs. Notably, plant PRRs appear as central components of multiprotein complexes at the plasma membrane that contain additional transmembrane and cytosolic kinases required for the initiation and specificity of immune signaling. PRR complexes are under tight control by protein phosphatases, E3 ligases, and other regulatory proteins, illustrating the exquisite and complex regulation of these molecular machines whose proper activation underlines a crucial layer of plant immunity.
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Plant Pathogenomics
April 28, 2014 4:18 AM
|
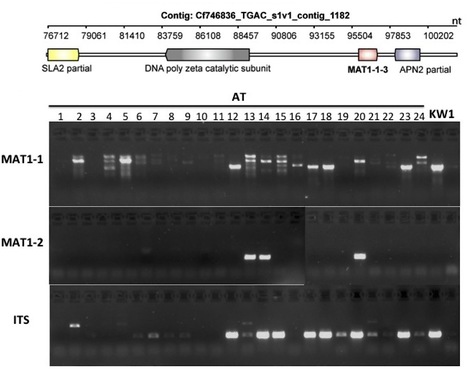
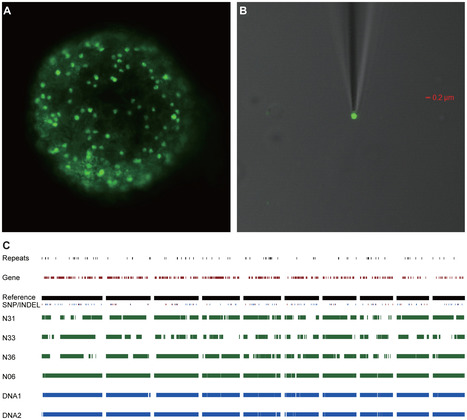
Ash dieback is a fungal disease of ash trees caused by Hymenoscyphus pseudoalbidus that has swept across Europe in the last two decades and is a significant threat to the ash population. This emergent pathogen has been relatively poorly studied and little is known about its genetic make-up. In response to the arrival of this dangerous pathogen in the UK we took the unusual step of providing an open access database and initial sequence datasets to the scientific community for analysis prior to performing an analysis of our own. Our goal was to crowdsource genomic and other analyses and create a community analysing this pathogen. In this report on the evolution of the community and data and analysis obtained in the first year of this activity, we describe the nature and the volume of the contributions and reveal some preliminary insights into the genome and biology of H. pseudoalbidus that emerged. In particular our nascent community generated a first-pass genome assembly containing abundant collapsed AT-rich repeats indicating a typically complex genome structure. Our open science and crowdsourcing effort has brought a wealth of new knowledge about this emergent pathogen within a short time-frame. Our community endeavour highlights the positive impact that open, collaborative approaches can have on fast, responsive modern science.
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Plants and Microbes
April 18, 2014 4:30 AM
|
Cytoplasmic plant immune receptors recognize specific pathogen effector proteins and initiate effector-triggered immunity. In Arabidopsis, the immune receptors RPS4 and RRS1 are both required to activate defense to three different pathogens. We show that RPS4 and RRS1 physically associate. Crystal structures of the N-terminal Toll–interleukin-1 receptor/resistance (TIR) domains of RPS4 and RRS1, individually and as a heterodimeric complex (respectively at 2.05, 1.75, and 2.65 angstrom resolution), reveal a conserved TIR/TIR interaction interface. We show that TIR domain heterodimerization is required to form a functional RRS1/RPS4 effector recognition complex. The RPS4 TIR domain activates effector-independent defense, which is inhibited by the RRS1 TIR domain through the heterodimerization interface. Thus, RPS4 and RRS1 function as a receptor complex in which the two components play distinct roles in recognition and signaling.
See also Perspective by Nishimura and Dangl http://www.sciencemag.org/content/344/6181/267.short
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Publications
March 31, 2014 3:51 AM
|
Both plants and animals rely on nucleotide-binding domain and leucine-rich repeat-containing proteins (NB-LRRs or NLRs) to respond to invading pathogens and activate immune responses. How plant NB-LRR proteins respond to pathogens is poorly understood. We undertook a gain-of-function random mutagenesis screen of the potato NB-LRR immune receptor R3a to study how this protein responds to the effector protein AVR3a from the oomycete pathogen Phytophthora infestans. R3a response can be extended to the stealthy AVR3aEM isoform of the effector while retaining recognition of AVR3aKI. Each one of 8 single amino acid mutations is sufficient to expand the R3a response to AVR3aEM and other AVR3a variants. These mutations occur across the R3a protein, from the N-terminus to different regions of the LRR domain. Further characterization of these R3a mutants revealed that at least one of them was sensitized, exhibiting a stronger response than the wild-type R3a protein to AVR3aKI. Remarkably, the N336Y mutation, near the R3a nucleotide-binding pocket, conferred response to the effector protein PcAVR3a4 from the vegetable pathogen Phytophthora capsici. This work contributes to understanding how NB-LRR receptor specificity can be modulated. Together with knowledge of pathogen effector diversity, this strategy can be exploited to develop synthetic immune receptors.
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Plants and Microbes
March 16, 2014 5:24 PM
|
Innate immunity relies on the perception of pathogen-associated molecular patterns (PAMPs) by pattern-recognition receptors (PRRs) located on the host cell’s surface. Many plant PRRs are kinases. Here, we report that the Arabidopsis receptor kinase EF-TU RECEPTOR EFR, which perceives the elf18 peptide derived from bacterial elongation factor Tu, is activated upon ligand binding by phosphorylation on its tyrosine residues. Phosphorylation of a single tyrosine residue, Y836, is required for activation of EFR and downstream immunity to the phytopathogenic bacterium Pseudomonas syringae. A tyrosine phosphatase, HopAO1, secreted by P. syringae, reduces EFR phosphorylation and derails subsequent immune responses. Thus host and pathogen battle to take control of PRR tyrosine phosphorylation used to initiate anti-bacterial immunity.
Via Jim Alfano, Kamoun Lab @ TSL

|
Scooped by
The Sainsbury Lab
March 12, 2014 6:00 PM
|

|
Scooped by
The Sainsbury Lab
March 3, 2014 5:50 AM
|

|
Rescooped by
The Sainsbury Lab
from Publications
February 27, 2014 7:34 AM
|
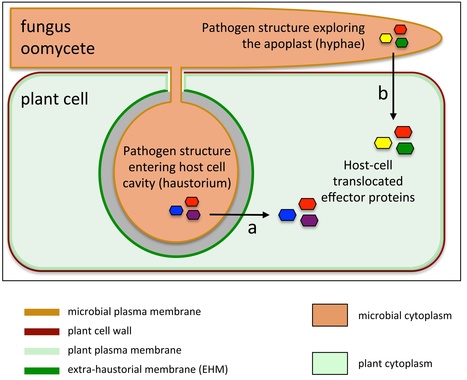
Fungal and oomycete plant parasites are among the most devastating pathogens of food crops. These microbes secrete effector proteins inside plant cells to manipulate host processes and facilitate colonization. How these effectors reach the host cytoplasm remains an unclear and debated area of plant research. In this article, we examine recent conflicting findings that have generated discussion in the field. We also highlight promising approaches based on studies of both parasite and host during infection. Ultimately, this knowledge may inform future broad spectrum strategies for protecting crops from such pathogens.
Via Kamoun Lab @ TSL

|
Scooped by
The Sainsbury Lab
February 17, 2014 4:15 AM
|

|
Scooped by
The Sainsbury Lab
January 15, 2014 10:18 AM
|
PLOS Genetics is an open-access

|
Scooped by
The Sainsbury Lab
January 2, 2014 8:10 AM
|
|

|
Rescooped by
The Sainsbury Lab
from Plants and Microbes
May 13, 2014 3:49 AM
|
Filamentous pathogens such as the oomycete Phytophthora infestans infect plants by developing specialized structures termed haustoria inside the host cells. Haustoria are thought to enable secretion of effector proteins into the plant cells. Haustorium biogenesis is therefore critical for pathogen accommodation in the host tissue. Haustoria are enveloped by a specialized host-derived membrane, the extrahaustorial membrane (EHM), which is distinct from the plant plasma membrane. The mechanisms underlying the biogenesis of the EHM are unknown. Remarkably, several plasma membrane localised proteins are excluded from the EHM but the remorin REM1.3 accumulates around P. infestans haustoria. Here, we used overexpression, co-localization with reporter proteins, and super-resolution microscopy in cells infected by P. infestans to reveal discrete EHM domains labelled by REM1.3 and P. infestans effector AVRblb2. Moreover, SYT1 synaptotagmin, another previously identified perihaustorial protein, localized to subdomains which are mainly not labelled by REM1.3 and AVRblb2. Functional characterization of REM1.3 revealed that it is a susceptibility factor that promotes infection by P. infestans. This activity, and REM1.3 recruitment to the EHM, require REM1.3 membrane binding domain. Our results implicate REM1.3 membrane micro-domains in plant susceptibility to an oomycete pathogen.
Via Kamoun Lab @ TSL

|
Scooped by
The Sainsbury Lab
May 7, 2014 8:05 AM
|

|
Scooped by
The Sainsbury Lab
April 28, 2014 4:20 AM
|

|
Rescooped by
The Sainsbury Lab
from Plant Pathogenomics
April 25, 2014 4:16 AM
|
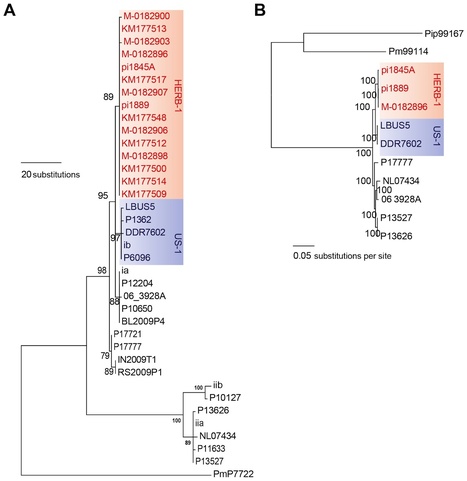
Since the dawn of agriculture, plant pathogens and pests have been a scourge of humanity. Yet we have come a long way since the Romans attempted to mitigate the effects of plant disease by worshipping and honoring the god Robigus. Books in the Middle Ages by Islamic and European scholars described various plant diseases and even proposed particular disease management strategies. Surprisingly, the causes of plant diseases remained a matter of debate over a long period. It took Henri-Louis Duhamel du Monceau's elegant demonstration in his 1728 “Explication Physique” paper that a “contagious” fungus was responsible for a saffron crocus disease to usher in an era of documented scientific inquiry. Confusion and debate about the exact nature of the causal agents of plant diseases continued until the 19th century, which not only saw the first detailed analyses of plant pathogens but also provided much-needed insight into the mechanisms of plant disease. An example of this is Anton de Bary's demonstration that a “fungus” is a cause, not a consequence, of plant disease. This coming of age of plant pathology was timely. In the 19th century, severe plant disease epidemics hit Europe and caused economic and social upheaval. These epidemics were not only widely covered in the press but also recognized as serious political issues by governments. Many of the diseases, including late blight of potato, powdery and downy mildew of grapevine, as well as phylloxera, were due to exotic introductions from the Americas and elsewhere. These and subsequent epidemics motivated scientific investigations into crop breeding and plant disease management that developed into modern plant pathology science over the 20th century. Nowadays, our understanding of plant pathogens and the diseases they cause greatly benefits from molecular genetics and genomics. All aspects of plant pathology, from population biology and epidemiology to mechanistic research, are impacted. The polymerase chain reaction (PCR) first enabled access to plant pathogen DNA sequences from historical specimens deposited in herbaria. Historical records in combination with herbarium specimens have turned out to provide powerful tools for understanding the course of past plant epidemics. Recently, thanks to new developments in DNA sequencing technology, it has become possible to reconstruct the genomes of plant pathogens in herbaria. In this article, we first summarize how whole genome analysis of ancient DNA has been recently used to reconstruct the 19th-century potato-blight epidemic that rapidly spread throughout Europe and triggered the Irish potato famine. We then discuss the exciting prospects offered by the emergence of the discipline of ancient plant pathogen genomics.
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Publications
April 7, 2014 3:49 AM
|
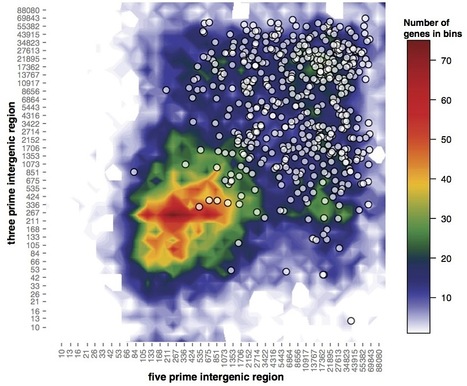
Genome architecture often reflects an organism’s lifestyle and can therefore provide insights into gene function, regulation, and adaptation. In several lineages of plant pathogenic fungi and oomycetes, characteristic repeat-rich and gene-sparse regions harbor pathogenicity-related genes such as effectors. In these pathogens, analysis of genome architecture has assisted the mining for novel candidate effector genes and investigations into patterns of gene regulation and evolution at the whole genome level. Here we describe a two-dimensional data binning method in R with a heatmap-style graphical output to facilitate analysis and visualization of whole genome architecture. The method is flexible, combining whole genome architecture heatmaps with scatter plots of the genomic environment of selected gene sets. This enables analysis of specific values associated with genes such as gene expression and sequence polymorphisms, according to genome architecture. This method enables the investigation of whole genome architecture and reveals local properties of genomic neighborhoods in a clear and concise manner.
Via Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Publications
March 28, 2014 5:55 AM
|
Rust fungi cause serious yield reductions on crops, including wheat, barley, soybean, coffee, and represent real threats to global food security. Of these fungi, the flax rust pathogen Melampsora lini has been developed extensively over the past 80 years as a model to understand the molecular mechanisms that underpin pathogenesis. During infection, M. lini secretes virulence effectors to promote disease. The number of these effectors, their function and their degree of conservation across rust fungal species is unknown. To assess this, we sequenced and assembled de novo the genome of M. lini isolate CH5 into 21,130 scaffolds spanning 189 Mbp (scaffold N50 of 31 kbp). Global analysis of the DNA sequence revealed that repetitive elements, primarily retrotransposons, make up at least 45% of the genome. Using ab initio predictions, transcriptome data and homology searches, we identified 16,271 putative protein-coding genes. An analysis pipeline was then implemented to predict the effector complement of M. lini and compare it to that of the poplar rust, wheat stem rust and wheat stripe rust pathogens to identify conserved and species-specific effector candidates. Previous knowledge of four cloned M. lini avirulence effector proteins and two basidiomycete effectors was used to optimise parameters of the effector prediction pipeline. Markov clustering based on sequence similarity was performed to group effector candidates from all four rust pathogens. Clusters containing at least one member from M. lini were further analysed and prioritized based on features including expression in isolated haustoria and infected leaf tissue and conservation across rust species. Herein, we describe 200 of 940 clusters that ranked highest on our priority list, representing 725 flax rust candidate effectors. Our findings on this important model rust species provide insight into how effectors of rust fungi are conserved across species and how they may act to promote infection on their hosts.
Via Francis Martin, Kamoun Lab @ TSL

|
Rescooped by
The Sainsbury Lab
from Publications
March 16, 2014 5:22 PM
|
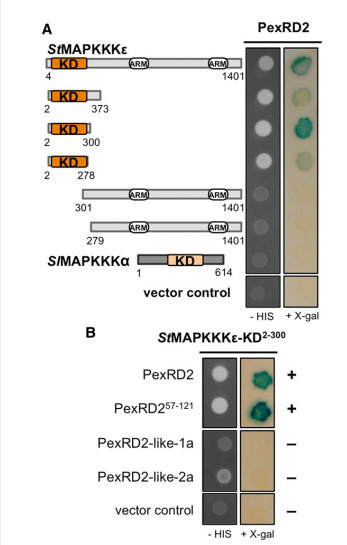
Mitogen-activated protein kinase cascades are key players in plant immune signaling pathways, transducing the perception of invading pathogens into effective defense responses. Plant pathogenic oomycetes, such as the Irish potato famine pathogen Phytophthora infestans, deliver RXLR effector proteins to plant cells to modulate host immune signaling and promote colonization. Our understanding of the molecular mechanisms by which these effectors act in plant cells is limited. Here, we report that the P. infestans RXLR effector PexRD2 interacts with the kinase domain of MAPKKKε, a positive regulator of cell death associated with plant immunity. Expression of PexRD2 or silencing MAPKKKε inNicotiana benthamiana enhances susceptibility to P. infestans. We show that PexRD2 perturbs signaling pathways triggered by or dependent on MAPKKKε. By contrast, homologs of PexRD2 from P. infestans had reduced or no interaction with MAPKKKε and did not promote disease susceptibility. Structure-led mutagenesis identified PexRD2 variants that do not interact with MAPKKKε and fail to support enhanced pathogen growth or perturb MAPKKKε signaling pathways. Our findings provide evidence that P. infestans RXLR effector PexRD2 has evolved to interact with a specific host MAPKKK to perturb plant immunity–related signaling.
Via Suayib Üstün, Kamoun Lab @ TSL

|
Scooped by
The Sainsbury Lab
March 12, 2014 4:23 PM
|

|
Scooped by
The Sainsbury Lab
March 3, 2014 5:31 AM
|

|
Scooped by
The Sainsbury Lab
February 21, 2014 6:13 AM
|

|
Rescooped by
The Sainsbury Lab
from Publications
January 31, 2014 3:41 AM
|
Accelerated gene evolution is a hallmark of pathogen adaptation following a host jump. Here, we describe the biochemical basis of adaptation and specialization of a plant pathogen effector after its colonization of a new host. Orthologous protease inhibitor effectors from the Irish potato famine pathogen, Phytophthora infestans, and its sister species, Phytophthora mirabilis, which is responsible for infection of Mirabilis jalapa, are adapted to protease targets unique to their respective host plants. Amino acid polymorphisms in both the inhibitors and their target proteases underpin this biochemical specialization. Our results link effector specialization to diversification and speciation of this plant pathogen.
Via Kamoun Lab @ TSL

|
Scooped by
The Sainsbury Lab
January 6, 2014 4:55 AM
|
PLOS Genetics is an open-access
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...










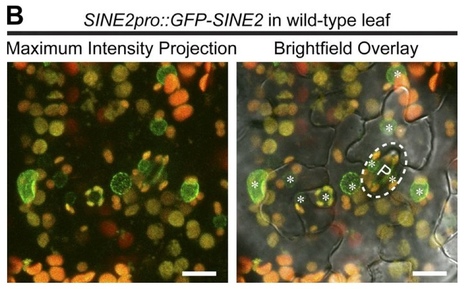





















Although a plethora of nuclear envelope (NE) transmembrane proteins (NETs) have been identified in opisthokonts, plant NETs are largely unknown. The only known NET homologues in plants are Sad1/UNC-84 (SUN) proteins, which bind Klarsicht/ANC-1/Syne-1 homology (KASH) proteins. Therefore, de novo identification of plant NETs is necessary. Based on similarities between opisthokont KASH proteins and the only known plant KASH proteins, WPP domain–interacting proteins, we used a computational method to identify the KASH subset of plant NETs. Ten potential plant KASH protein families were identified, and five candidates from four of these families were verified for their NE localization, depending on SUN domain interaction. Of those, Arabidopsis thaliana SINE1 is involved in actin-dependent nuclear positioning in guard cells, whereas its paralogue SINE2 contributes to innate immunity against an oomycete pathogen. This study dramatically expands our knowledge of plant KASH proteins and suggests that plants and opisthokonts have recruited different KASH proteins to perform NE regulatory functions.