 Your new post is loading...
 Your new post is loading...

|
Scooped by
The Sainsbury Lab
January 28, 5:46 AM
|
Pathogen pressure threatens legume crop productivity worldwide. Nucleotide-binding leucine-rich repeat (NLR) immune receptors serve as crucial plant resistance genes, recognizing pathogens and triggering immunity. However, the extent and patterns of NLR expression in different tissues and organs, notably across evolutionary time, remain largely uncharacterized. To investigate tissue-specificity of NLR expression in the Fabaceae (legumes), we conducted comparative analyses integrating phylogenomics and transcriptomics in root and shoot tissues across different legume species. The NLR repertoires of 28 legumes were grouped into five monophyletic clades: coiled-coil NLR (CC-NLR), Toll/interleukin-1 receptor NLR (TIR-NLR), G10-subclade CC NLR (CCG10-NLR), RESISTANCE TO POWDERY MILDEW 8-like CC NLR (CCR-NLR), and TIR-NB-ARC-like β-propeller WD40/tetratricopeptide repeats (TNPs). Most legume NLRs belonged to CC-NLR and TIR-NLR clades, followed by CCG10-NLR, CCR-NLR, and TNP clades. In seven of these species, comparative analysis of NLR expression in leaves versus roots revealed that over half (~57%) of expressed NLR genes showed predominant expression in one tissue: 34% in roots (451/1336), and 23% in leaves (311/1336). We identified 324 root-specific NLRs, 171 leaf-specific NLRs, and 841 non-specific NLRs, with an average tissue specificity per species of 32%. The closely related species grass pea (Lathyrus sativus) and pea (Pisum sativum) were an exception, showing higher levels of leaf-specific rather than root-specific NLR expression. We also identified conserved tissue expression patterns across legume species, resulting in a comprehensive resource describing tissue expression bias, enrichment, and specificity for 113 phylogenetic NLR subclasses. These legume NLR repertoires will support comparative studies between species and inform precision-breeding programs considering tissue expression patterns.

|
Scooped by
The Sainsbury Lab
January 15, 4:53 AM
|
Plants employ cell-surface and intracellular immune receptors to perceive pathogens and activate defense responses. Recent advances in mechanistic understanding of how cell-surface and intracellular immune receptors convert recognition of molecular patterns or effectors into defense activation, combined with the knowledge of receptor repertoire variation both within and between species, allow transfer of immune receptors between species to increase the spectrum of recognition specificities. Here, we summarize recent progress in the functional transfer of immune receptors within and between plant families. We also discuss challenges that limit the transferability of intracellular immune receptors, including the requirement of additional host factors or downstream components and their incompatibility between donor and recipient species. Finally, we provide an overview of future perspectives for bioengineering disease-resistant crops through immune receptor transfer.

|
Scooped by
The Sainsbury Lab
January 13, 12:24 PM
|
Plant V-ATPase serves as a primary active proton pump of the endomembrane system and is crucial for the stress response. However, the role of the C subunit of V-ATPase (VHA-C) in effector-triggered immunity remains poorly understood. Here, we reveal that Phytophthora infestans evolved a pair of RxLR effectors, AL3 and Avr2, which are expressed sequentially and both target the host VHA-C (StATP6V1C1) and StBSL1. In the early stage of P. infestans infection, AL3 promotes the assembly of StATP6V1C1 with subunits G and E, leading to increased V-ATPase activity and cytoplasmic acidification. Subsequently, Avr2 inhibits the StWNK10-catalyzed Ser-261 phosphorylation of StATP6V1C1, thereby retarding V-ATPase activity and causing intracellular alkalinization. In cultivars absence of two immune receptors, this pH shift facilitates the interactions of the two effectors with downstream susceptibility factors of StBSL1 at various stages of infection, which may promote the onset and development of the disease. As coping strategy, plants independently evolve two NLRs, R2 and Rpi-mcq1, guard both StATP6V1C1 and StBSLs to perceive effectors thereby mitigating the risk of late blight. Our findings establish a new arms race battlefield between plants and oomycetes, highlighting the role of intracellular pH homeostasis in both effector-triggered susceptibility (ETS) and effector-triggered immunity (ETI).

|
Scooped by
The Sainsbury Lab
January 5, 7:06 AM
|
The advent of single-cell genomic technologies has revolutionized plant cell biology by revealing cellular heterogeneity in plant tissues and organs with unprecedented resolution. Methods like single-cell transcriptomics, epigenomics, and multi-omics integration have deepend insights into molecular mechanisms governing plant development, function, and evolutionary adaptations (Xu and Jackson, 2025). Notably, these approaches have facilitated comprehensive cell atlases mapping gene expression, chromatin accessibility, and regulatory networks across species (Wang et al., 2025; Guo et al., 2025), identifying marker genes, differentiation trajectories, and spatiotemporal regulatory in processes like root development, and seed germination (Zhang et al., 2021; Yao et al., 2024). However, proliferating single-cell studies face a major bottleneck from inconsistent cell type annotations, which impede data integration and crossspecies comparison. A unified, phylogenetically informed atlas of plant cellular diversity is essential for elucidating molecular developmental and evolutionary principles (Sebé-Pedrós et al., 2025).

|
Scooped by
The Sainsbury Lab
December 30, 2025 7:09 AM
|
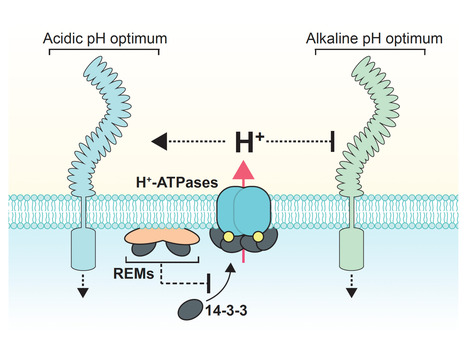
The power of hydrogen (pH) regulates virtually all cellular activities. In both plants and animals, cell-to-cell variations in pH correlate with key developmental transitions1–5, yet the underlying regulators and associated functions remain elusive. Here, we report that members of the REMORIN (REM) protein family function as inhibitors of the H+-ATPases thereby promoting extracellular pH (pHe) alkalinization. This, in turn, regulates various cell surface processes, including steroid hormone signaling, and coordinates developmental transitions in the Arabidopsis thaliana root. Inhibition of H+-ATPases by REMs represents an evolutionary innovation that predates the origin of the root system itself. This study thus uncovers an ancient alkalinization mechanism co-opted by the root developmental program and infers that pHe patterning may have shaped morphogenesis evolution.

|
Scooped by
The Sainsbury Lab
December 18, 2025 6:17 AM
|
- The morphology of starch granules is a major determinant of the functional and nutritional properties of starch and is highly variable among cereal species. Much of this morphological variation stems from differences in the spatial and temporal patterns of starch granule initiation in amyloplasts during grain development. Simple granules are thought to arise from a single initiation per amyloplast (e.g. in Brachypodium distachyon), whereas compound granules develop from multiple initiations per amyloplast (e.g. in rice).
- We used live-cell imaging to visualise amyloplasts in the developing endosperm of Brachypodium, using transgenic lines expressing a fluorescent amyloplast reporter.
- We discovered that the simple-type starch granules in Brachypodium can arise from multiple initiations per amyloplast. The amyloplasts showed dynamic changes in their structure and formed two types of stromules: stable stromules that formed a stromal continuum between amyloplasts, and short-lived stromules that were more dynamic. We also observed actin-dependent movement of amyloplasts within endosperm cells, and movement of starch granules within the amyloplasts.
- Our results suggest complex and pleiomorphic amyloplast organisation and mobility that could influence granule formation. This goes beyond the existing ‘one granule, one amyloplast’ model for simple-type granules and advances our understanding of both amyloplast biogenesis and granule formation.

|
Scooped by
The Sainsbury Lab
December 18, 2025 6:07 AM
|
Enterobacter cloacae is a Gram-negative nosocomial human pathogen that inhabits diverse ecological niches. Its genome encodes a conserved set of putative chitin-active enzymes, including a lytic polysaccharide monooxygenase (LPMO), termed EcLPMO, which we functionally characterized in this study. EcLPMO is a tetra-modular protein consisting of an auxiliary activity family 10 (AA10) catalytic domain, two central domains of unknown function (DUF-A and DUF-B), and a C-terminal carbohydrate-binding module (CBM73). Functional assays using full-length EcLPMO and its truncated variants demonstrated that the AA10 domain oxidatively cleaves chitin at the C1 position. The CBM73 module enhances chitin binding and promotes synergy with endogenous chitinases. EcLPMO exhibited synergy with the unimodular chitinase EcChiA, resulting in up to 14-fold and 60-fold increase in GlcNAc release from α- and β-chitin, respectively. Deletion of both DUFs reduced EcLPMO activity. While DUF-A alone and the association of DUF-A and DUF-B showed limited chitin binding, DUF-B alone exhibited no binding, suggesting a distinct role. Using AlphaFold3, we observed that the DUF-B domain contains two highly conserved histidines that coordinate the AA10-bound copper, forming a previously unreported 'inter-domain tetra-histidine copper coordination' center. These findings highlight the structural and functional complexity of EcLPMO and suggest that its accessory domains, particularly DUF-B, may contribute to enzyme stability and substrate interaction. We speculate that DUF-B may protect the LPMO active site from oxidative damage, a feature that could prove crucial in its ecological and pathogenic contexts.

|
Scooped by
The Sainsbury Lab
December 16, 2025 9:06 AM
|
Following the perception of pathogen virulence proteins in plants, nucleotide-binding and leucine-rich repeat immune receptors (NLRs) are activated via a wide range of mechanisms. Singleton NLRs can both perceive effectors and trigger an immune response, whereas other NLRs specialise in either pathogen recognition (sensor NLRs) or activation of the immune response (helper NLRs). Sensor and helper NLRs can function as genetically linked pairs or in unlinked receptor networks. Although growing evidence suggests that NLRs conditionally oligomerise upon activation, our understanding of the resting state of NLRs prior to effector perception remains limited. Here, we investigated the oligomeric state of the genetically linked rice (Oryza sativa) sensor Pik-1 and helper Pik-2 NLR pair prior to effector activation when transiently expressed in Nicotiana benthamiana leaves. We show that both wild-type Pikm-1 and engineered Pikm-1Enhancer sensors associate with Pikm-2 and form ∼1 MDa hetero-complexes in the resting state that accumulate at the plasma membrane. Our findings contribute to the growing evidence that pre-activation mechanisms vary widely across NLRs. This knowledge could be leveraged for disease resistance engineering strategies complementary to approaches focussing solely on effector binding.

|
Scooped by
The Sainsbury Lab
December 16, 2025 9:03 AM
|
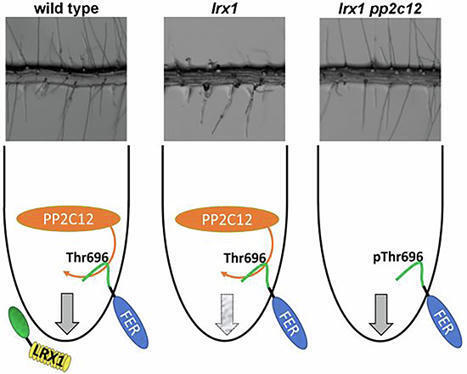
Plants have evolved an elaborate cell wall integrity (CWI) sensing system to monitor and modify cell wall formation. LRR-extensins (LRXs) are cell wall-anchored proteins that bind RAPID ALKALINIZATION FACTOR (RALF) peptide hormones and induce compaction of cell wall structures. LRXs also form a signaling platform with RALFs and the transmembrane receptor kinase FERONIA (FER) to maintain cell wall integrity. LRX1 of Arabidopsis thaliana is predominantly expressed in root hairs, and lrx1 mutants develop defective root hairs. Here, we identify a regulator of LRX1-RALF-FER signaling as a suppressor of the lrx1 root hair phenotype. The repressor of lrx1_23 (rol23) gene encodes PP2C12, a clade H phosphatase that interacts with FER and dephosphorylates Thr696 in the FER activation loop in vitro. Expression of FER phospho-mimetic and phospho-null mutants in an lrx1 fer-4 background demonstrates that phosphorylation of FER at Thr696 is essential for suppression of lrx1 phenotypes by rol23. The LRX1-related function of PP2Cs appears clade H-specific. Collectively, our data suggest that LRX1 acts upstream of the RALF-FER signaling module and that PP2C12 inhibits FER via activation-loop dephosphorylation.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:55 AM
|
The wheat race-specific resistance gene Lr14a encodes an ankyrin-repeat transmembrane domain protein that confers immunity against the fungal pathogen Puccinia triticina, but the underlying mechanisms remain unclear. Here, using a range of biological systems and methods, we show that Lr14a functions as a Ca²⁺-permeable channel crucial for leaf rust resistance in wheat. Interestingly, we also found that the pathogen-inducible expression of Lr14a triggers guard cell hypersensitive response (HR) non-cell autonomously as a mechanism to control pathogen entry. Our results suggest that Lr14a regulates stomatal immunity non-cell autonomously through the mediation of calcium signaling.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:48 AM
|
Plants encounter diverse pathogens and have evolved a two-layered innate immune system to detect pathogen molecules and activate defense mechanisms that restrict infection. Most cloned plant Resistance (R) genes encode NLR immune receptors. NLR genes are often found in clusters of paralogs with sequence and copy number variation; whether these NLR clusters evolve in response to single or multiple pathogens has been unclear. We report here the isolation of a Phytophthora capsici resistance gene, Rpc2, along with a novel P. infestans resistance gene, Rpi-amr5, from two Solanum americanum accessions. These orthologous genes reside in the Rpi-amr1 cluster, which has previously been associated with resistance to P. infestans. By screening RXLR effector libraries of P. infestans and P. capsici, we identified multiple effectors recognized by both NLRs. Our findings highlight the complexity of NLR clusters and evolution driven by interactions with multiple pathogens. This work will underpin efforts to elevate resistance against Phytophthora pathogens and enhances our understanding of NLR evolution.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:45 AM
|
Nucleotide-binding and leucine-rich repeat immune receptors of the NRC clade act in one direction, with autoactive helpers able to trigger hypersensitive cell death without their sensor partners.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:39 AM
|
Precise ligand recognition by closely related leucine-rich repeat receptor kinases (LRR-RKs) is essential for plants to coordinate immunity, development, and environmental adaptation. Here, we show how HAESA-LIKE 3 (HSL3) specifically recognizes the folded, disulfide-stabilized CTNIP/SCREW phytocytokines in Arabidopsis. Quantitative binding assays define a minimal CTNIP4 region required for high-affinity HSL3 interaction and signaling activation. A 2.12 Å crystal structure of the HSL3-CTNIP4 complex reveals a unique C-terminal receptor pocket that accommodates the peptide’s cyclic architecture through a combination of hydrophobic and polar contacts, a feature absent in the closely related HAE/HSL LRR-RKs. The cyclic CTNIP4 fold further establishes a largely hydrophobic interface that bridges HSL3 to the SERK co-receptor, forming a distinct activation surface. Together, these structural, biochemical and physiological insights uncover a previously unrecognised mechanism of peptide perception and receptor activation, highlighting how subtle architectural variations enable precise ligand selectivity among highly conserved plant receptor kinases.
|

|
Scooped by
The Sainsbury Lab
January 19, 6:22 AM
|
Genetically encoded pigments are powerful visual reporters and creative tools for biology, yet in plants the palette of pigment biosynthesis genes has remained largely limited to red betacyanins encoded by RUBY. Here we develop and characterize three new polycistronic constructs AMBER_v1, AMBER_v2, and GOLD that contain betalain biosynthesis enzymes to produce yellow, fluorescent betaxanthins in plant tissues. These tools expand the palette of publicly available pigmentation genes for use in plant research, education, and floral design.

|
Scooped by
The Sainsbury Lab
January 14, 4:42 AM
|
The receptor-like cytoplasmic kinase BIK1 and its close homologue PBL1 have been widely recognized as central components of plant immunity. However, most genetic studies of BIK1 and PBL1 functions were carried out with single transfer DNA (T-DNA) insertional mutant alleles. Some phenotypes observed in these mutants, for example autoimmunity, have been difficult to reconcile with the proposed role of BIK1 and PBL1 in pattern-triggered immunity. In this study, we generated several new alleles of bik1 and pbl1 by CRISPR–Cas9-based gene editing and systematically analysed these mutants alongside existing T-DNA insertional lines. These analyses reinforced the central role of BIK1 and PBL1 in pattern-triggered immunity mediated by both receptor kinases and receptor-like proteins. At the same time, however, we revealed several pleiotropic phenotypes associated with T-DNA insertions that are not necessarily linked to loss of BIK1 or PBL1 function. Further analyses of newly generated bik1 pbl1 double mutants uncovered an even greater contribution of these kinases to immune signalling and disease resistance than previously appreciated. These findings clarify longstanding ambiguities surrounding BIK1 and PBL1 functions. Thorough genetic analyses by four laboratories reinforce the central role of the protein kinase BIK1 in plant immunity. Other unrelated functions proposed for BIK1, however, are associated with genome rearrangement in a single transfer DNA mutant.

|
Scooped by
The Sainsbury Lab
January 8, 6:57 AM
|
Effectors are pathogen proteins that facilitate infection by manipulating plant immunity. Computational programs have been developed that identify effectors from sequence data. Many of these programs use internal models that have unavoidable biases due to their training processes and the diverse nature of effector sequence, function and phylogeny. Each programs ability to predict effectors across a broad range of plant pathogens is therefore limited. We hypothesised that a meta-predictor constructed using machine learning (ML) approaches could integrate predictions from multiple programs and improve our ability to predict effectors more accurately from bacteria, fungi, and oomycetes. We trained a range of classifiers using classical ML approaches and deep neural networks (DNNs), then selected eight: Random Forest (RF), Support Vector Machine (SVM), Extreme Gradient Boosting (XGBoost), and five DNNs for evaluation. The training, test and validation data were carefully curated from effector and non-effector annotated sequence derived from the training and sample data of six programs: EffectorP 3.0, deepredeff, WideEffHunter, EffectorO, EffectiveT3, T3SEpp. The models were tested against existing programs on a test dataset, and we observed better performance from our models. The best-performing model was a DNN (Model_2) that balanced improved sensitivity with specificity across the three taxa. We observed using SHAP that all the features contributed to the output of Model_2, which might be the reason for its superior performance. The DNN was developed into a package, fimep, to allow easy use of our model.

|
Scooped by
The Sainsbury Lab
December 30, 2025 11:46 AM
|
Nucleotide-binding leucine-rich repeat (NLR) immune receptors sense pathogen molecules and oligomerize, initiating defense signaling. Some NLRs function poorly at elevated temperatures for unknown reasons. We show that temperature-sensitive NLRs retain ligand binding at elevated temperatures but are impaired in oligomerization. We identify key residues involved in temperature resilience. Structural modeling reveals stabilizing intramolecular interactions of the NB-ARC domain with surface residues of the adjacent leucine-rich repeat (LRR) that preserve receptor integrity and functionality under heat stress. These insights enable in silico classification of NLRs as temperature-sensitive or -tolerant and underpin design of temperature tolerant variants of temperature sensitive NLRs. These findings provide a mechanistic basis for temperature sensitivity in plant immune receptors and enable engineering of temperature-tolerant disease resistance in crops.

|
Scooped by
The Sainsbury Lab
December 30, 2025 6:16 AM
|
Diploid potato breeding enables faster genetic improvement via selection against deleterious alleles in inbred lines, unlike breeding by intercrossing tetraploid varieties. Starch is the major source of calories in potato tubers, but the starch properties of diploid lines have rarely been reported. In this study, we provide a comprehensive characterisation of tuber and starch properties in two diploid lines that are early isolates from the Solynta breeding program, B26 and B100, and their F1 hybrids. B100 produced fewer, but larger tubers compared to B26, and both diploid lines produced tubers that are smaller than the tetraploid variety, Clearwater Russet. The low tuber yield of B100 correlates with its high self-compatibility and fruit production. Pruning of fruits in B100 significantly increased total tuber yield per plant by stimulating more tuber initiations, but had no effect on average tuber weight, starch content or starch structure. Among the diploid, hybrid and tetraploid lines examined, there were no differences in the total starch content of tubers. Although amylopectin structure and amylose content were similar between the two diploid lines and the tetraploid comparison, B26 had elevated levels of resistant starch and a striking elongated granule morphology. Our results showcase the variation in source-sink relations and starch structure in diploid potato breeding material, demonstrating their potential for research into the genetics underpinning metabolic and quality traits.

|
Scooped by
The Sainsbury Lab
December 18, 2025 6:08 AM
|
Subjects Bacterial techniques and applicationsFungal biologySymbiosis Eukaryotes often host prokaryotic cells inside their own cells. Hosts of such ‘endosymbionts’ can rely on these organisms for metabolism or the production of other necessary molecules.

|
Scooped by
The Sainsbury Lab
December 16, 2025 9:09 AM
|
Angiosperm seed formation requires the coordinated development of the products of double fertilization, the embryo and the endosperm. The endosperm mediates efficient nutrient transfer from surrounding maternal tissues to the developing embryo. This function requires a polarized tissue organization, which manifests as early polar gene expression and polar cellularization dynamics. We show that the receptor kinase HAIKU2 acts in coordination with the transcription factor WRKY10/MINISEED3 to ensure robust endosperm polarity establishment through the activity of the homeodomain transcription factors WUSCHEL-RELATED HOMEOBOX 8 and 9. This process depends on egg cell fertilization and is mediated through the peptide PATHOGEN-INDUCED PEPTIDE-LIKE 7, which acts as a HAIKU2 ligand. Our results reveal how a molecular paracrine dialogue between the embryo and endosperm ensures optimal seed developmental coordination.

|
Scooped by
The Sainsbury Lab
December 16, 2025 9:05 AM
|
Lichens are symbiotic associations between filamentous fungi and photosynthetic micro-organisms, such as algae or cyanobacteria, that result in a single anatomically-complex structure that can thrive in environments inhospitable to most organisms, including arctic tundra, high mountains, and deserts. Recent evidence suggests that lichens may be even more complex than previously appreciated, containing multiple microbial constituents that operate as mini-ecosystems, but how genomes of the principal fungal symbiont (which provides the majority of biomass in lichen tissue) have been shaped during evolution is largely unexplored. Recently, giant transposable elements called Starships have been found in many genomes of filamentous fungi, but to which extent they occur in lichen-forming fungi is not known. In this report, we describe a Starship-like element from the lichen fungus Xanthoria parietina. This element, named Tangerine, contains several genes that have signatures of horizontal gene transfer from non-lichen-forming fungi, most likely from black yeasts of the Chaetothyriales, that are often lichen-associated. Furthermore, the “captain” gene responsible for transposition of the Starship defines a small lichen-specific clade of tyrosine recombinases within clade 1 of the tyrosine recombinase typology, suggesting a longstanding association of these elements with lichen-forming fungi. Internal repeats within Tangerine, and other sites in Xanthoria genomes, are affected by repeat-induced point mutation (RIP), a mechanism of genome defense against transposable elements, consistent with fungal sexual reproduction which always precedes new lichen formation by Xanthoria. We conclude that Starships may have played a significant, yet hitherto unrecognized role, in lichen genome evolution.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:58 AM
|
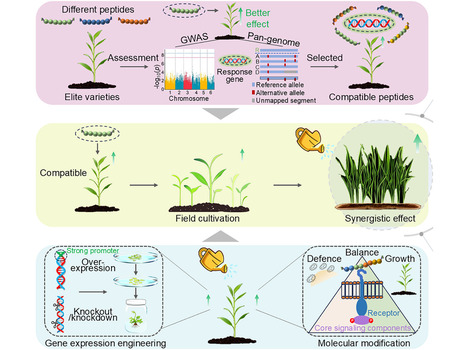
Global agriculture faces critical challenges due to the overreliance on chemical pesticides, driving an urgent need for eco-friendly biopesticides and biostimulants (BioP&S). Plant-derived peptides, evolved as natural regulators of growth, development, and stress adaptation, offer immense potential as biodegradable and biocompatible alternatives. However, their commercialization remains constrained by limited exploration of the diversity and activity, high production costs, incomplete ecological risk evaluations, and undefined application scenarios. This Perspective overviews emerging discoveries and proposes integrated frameworks for plant peptide identification, molecular design, biomanufacturing, and ecological impact assessments integrated with germplasm development and field application systems. To overcome existing bottlenecks, we discuss the integrative potential of emerging technologies that synergistically combine artificial intelligence for high-throughput peptide discovery and de novo structural refinement, nanotechnology for enhancing environmental resilience and targeted delivery, and synthetic biology for developing industrial biomanufacturing platforms. We emphasize the need to align phytopeptide BioP&S with compatible germplasm resources, stage-specific crop requirements, and complementary chemical pesticides to maximize their efficacy, cost-effectiveness, and trait-specific agronomic performance by integrating with precision agriculture systems. Future advancements will rely on interdisciplinary innovations and policy support to unlock their full potential in enhancing crop resilience, productivity, and quality while ensuring ecological sustainability.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:49 AM
|
Lichens are symbiotic assemblies consisting of multiple organisms, chiefly a fungus and a photosynthetic microorganism, or photobiont. Among diverse photobionts, the most prevalent is the chlorophyte alga Trebouxia. We produced a chromosome-level assembly of Trebouxia sp. ‘A48’, a photobiont of Xanthoria parietina. The genome was assembled into 20 contigs, of which 16 had telomeric repeats at both ends and likely represent complete chromosomes. We compared this genome with those of other Trebouxia species and analyzed it to investigate adaptations to the lichen lifestyle. We then used the genome to profile gene expression in axenic culture and in lichen thalli. The predicted secretome is enriched in hydrolases and redox enzymes and contains carbohydrate-binding proteins potentially involved in cell-to-cell recognition and adhesion. We identified genes potentially involved in carbon concentrating and confirmed two instances of ancient horizontal gene transfer from fungi. The genome and the strain of Trebouxia sp. ‘A48’ provide a resource for the community to research algal evolution and lichen symbiosis.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:46 AM
|
Intracellular immune receptors protect plants from microbial invasion by detecting and responding to pathogen-derived effector molecules, often triggering cell death responses. However, pathogen effectors can evolve to avoid immune recognition, resulting in devastating diseases that threaten global agriculture. Here, we show that an integrated Exo70 domain from the barley NLR RGH2 can interact with both the rice blast pathogen effector AVR-Pii and a closely related wheat blast variant. We used structure-led engineering to develop RGH2+ that shows increased binding affinity towards AVR-Pii variants and increased cell death responses on heterologous expression in Nicotiana benthamiana. Infection assays in transgenic barley lines carrying RGH2+ with the paired NLR RGH3 indicate a reduced susceptibility to blast strains expressing AVR-Pii variants. These results demonstrate the potential of engineering NLR receptors as an effective strategy for improving resistance towards one of the most destructive diseases affecting cereal production.

|
Scooped by
The Sainsbury Lab
December 16, 2025 8:43 AM
|
Plant science education needs urgent attention. Skilled plant scientists are needed to address major environmental and societal challenges, and global communities require plant-aware professionals to drive impactful policy, research and environmental stewardship. This manifesto was collaboratively generated by a community of educators who gathered to reflect on the state of plant science education. The forward-facing document provides a clear strategy for plant science education, complementing existing research strategies. Five themes were identified as essential for meeting the evolving needs of plant science, educators and learners: (i) plants must be at the centre of an education that addresses global challenges and societal values; (ii) plant science education must prepare students for their futures using bold and effective pedagogies; (iii) equity, diversity and inclusion must be robustly embedded in educational practices; (iv) local and strategic partnerships (with industry and beyond) are required to strengthen academic education; and (v) plant science educators need resources and opportunities to develop and connect. The manifesto is intended as a framework for change. Educators, funders, publishers, industry representatives, policymakers and all other members of our communities must commit to sustained investment in plant science education. By proactively and collectively embracing the recommendations provided, the sector has an opportunity to cultivate a new generation equipped with the knowledge, skills and passion to unlock the full potential of photosynthetic organisms.
|

 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...
































The availability of draft crop plant genomes allows the prediction of the full complement of genes that encode NB-LRR resistance gene homologs, enabling a more targeted breeding for disease resistance. Recently, we developed the RenSeq method to reannotate the full NB-LRR gene complement in potato and to identify novel sequences that were not picked up by the automated gene prediction software. Here, we established RenSeq on the reference genome of tomato (Solanum lycopersicum) Heinz 1706, using 260 previously identified NB-LRR genes in an updated Solanaceae RenSeq bait library.
Result: Using 250-bp MiSeq reads after RenSeq on genomic DNA of Heinz 1706, we identified 105 novel NB-LRR sequences. Reannotation included the splitting of gene models, combination of partial genes to a longer sequence and closing of assembly gaps. Within the draft S. pimpinellifolium LA1589 genome, RenSeq enabled the annotation of 355 NB-LRR genes. The majority of these are however fragmented, with 5[prime]- and 3[prime]-end located on the edges of separate contigs. Phylogenetic analyses show a high conservation of all NB-LRR classes between Heinz 1706, LA1589 and the potato clone DM, suggesting that all sub-families were already present in the last common ancestor. A phylogenetic comparison to the Arabidopsis thaliana NB-LRR complement verifies the high conservation of the more ancient CCRPW8-type NB-LRRs. Use of RenSeq on cDNA from uninfected and late blight-infected tomato leaves allows the avoidance of sequence analysis of non-expressed paralogues.