Your new post is loading...
 Your new post is loading...

|
Scooped by
The Sainsbury Lab
July 16, 10:54 AM
|
The rice blast fungus Magnaporthe oryzae secretes a battery of effector proteins to facilitate host infection. Among these effectors, pathogenicity toward weeping lovegrass 2 (Pwl2) was originally identified as a host specificity determinant for the infection of weeping lovegrass (Eragrostis curvula) and is also recognized by the barley (Hordeum vulgare) Mla3 resistance protein. However, the biological activity of Pwl2 remains unknown. Here, we showed that the Pmk1 MAP kinase regulates PWL2 expression during the cell-to-cell movement of M. oryzae at plasmodesmata-containing pit fields. Consistent with this finding, we provided evidence that Pwl2 binds to the barley heavy metal–binding isoprenylated protein HIPP43, which results in HIPP43 displacement from plasmodesmata. Transgenic barley lines overexpressing PWL2 or HIPP43 exhibit attenuated immune responses and increased disease susceptibility. In contrast, a Pwl2SNDEYWY variant that does not interact with HIPP43 fails to alter the plasmodesmata localization of HIPP43. Targeted deletion of 3 PWL2 copies in M. oryzae resulted in a Δpwl2 mutant showing gain of virulence toward weeping lovegrass and barley Mla3 lines, but reduced blast disease severity on susceptible host plants. Taken together, our results provide evidence that Pwl2 is a virulence factor that suppresses host immunity by perturbing the plasmodesmatal deployment of HIPP43.

|
Scooped by
The Sainsbury Lab
July 16, 5:55 AM
|
The Arabidopsis TIR-NLR immune receptors RPS4 and RRS1 function together to enable recognition of multiple effector proteins including AvrRps4 and PopP2. We show here that both in the presence and absence of effector, RPS4 and RRS1 form an oligomer that does not change in size upon effector provision. Oligomer formation involves interactions between the RPS4 and RRS1 TIR domains and requires nucleotide binding capacity in RPS4. RPS4 mutants that lose TIR domain NADase activity abrogate immune activation but retain oligomerization. A cysteine residue in the RPS4 LRR domain contributes to oligomer stabilization. We propose that upon effector recognition, conformational changes in the complex relieve inhibition of RPS4 TIR domains by RRS1 TIR domains, enabling proximity between RPS4 TIR domains to create NADase activity.

|
Scooped by
The Sainsbury Lab
July 16, 5:53 AM
|
Dioecious plants, which have distinct male and female individuals, constitute ∼5% of angiosperm species and have emerged frequently and independently from hermaphroditic ancestors. Although recent molecular studies of sex determination have started to reveal the diversity of the genetic systems underlying dioecy, research on the evolution of dioecy is limited, especially in monocots. Here, we describe the molecular basis of sex determination in the monocot Dioscorea tokoro, a dioecious wild yam endemic to East Asia. Chromosome-scale and haplotype-resolved genome assemblies and linkage analysis suggested that this plant has a male heterogametic sex determination (XY) system, with sex determination regions located on chromosome 3. Sequence read coverage analysis of the sex chromosomes revealed X- and Y-specific regions in putative pericentromeric chromosome regions. Within the Y-specific region, we identified two candidate genes that are likely involved in sex determination: BLH9, encoding a homeobox protein, and HSP90, encoding a molecular chaperone. BLH9 has similar functions to AtBLH9 in Arabidopsis thaliana. BLH9 is thought to suppress female organ development, whereas HSP90 might be required for pollen development. These results shed light on the complex evolution of dioecy in plants.

|
Scooped by
The Sainsbury Lab
July 16, 5:48 AM
|
Plants constantly monitor their environment to adapt to potential threats to their health and fitness. This involves cell-surface receptors that can detect conserved microbe-associated molecular patterns (MAMPs) or endogenous immunogenic signals, initiating signaling pathways to induce broad-spectrum disease resistance, known as pattern-triggered immunity (PTI). In Arabidopsis thaliana, the leucine-rich repeat receptor kinase (LRR-RK) MIK2 is an exceptionally versatile receptor involved in the perception of the vast family of Brassicales-specific endogenous SCOOP peptides as well as potential MAMPs derived from Fusarium and related fungi. Although only plant species belonging to the order of Brassicales encode genes for SCOOP peptides and show SCOOP-responsiveness, the Fusarium-derived elicitor fraction also induces PTI responses in plants from other lineages. In this study, we demonstrate that Fusarium elicitor-responsiveness and proteins belonging to the MIK2-clade are widely conserved among seed plants. We identified a MIK2-clade protein from tomato, which shares properties of AtMIK2 in the perception of the Fusarium elicitor but not of SCOOP peptides. Tomato mutants lacking the receptor show compromised PTI responses to the fungal elicitor and enhanced susceptibility to infection by Fusarium oxysporum. Our data provide insights into the evolutionary trajectory of MIK2 as a multifunctional receptor involved in plant immunity.

|
Scooped by
The Sainsbury Lab
July 16, 5:43 AM
|
Our usual encounters with fungi are when we observe mushrooms in forests or moulds on food that we failed to eat on time. In both instances, however, we are only seeing a very limited view of fungal growth. Fungi are osmotrophs, which means that they consume the food that surrounds them by secreting enzymes to degrade polymers into simple sugars, fatty acids and amino acids. This type of growth has a number of consequences - it explains why fungi secrete toxins and antibiotics to protect their food sources from competitors and why they can grow so rapidly. It also explains why fungi have evolved the capacity to forcefully invade hard substrates, such as wood, animal skins and the cuticles of plants. By doing so, they can access new sources of food, often inaccessible to their competition, and this has enabled fungi to become highly successful pathogens of both animals and plants, causing diseases in organisms as diverse as insects, amphibians, humans, reptiles, and rice plants. It is becoming clear that, in addition to their prodigious secretion of enzymes to degrade complex substrates, fungi can exert very substantial physical forces. Such forces are probably essential for many aspects of the fungal lifestyle, including colonisation of their usual habitats like soil and leaf litter, which require penetration and invasion to enable their digestion by fungi. But for pathogenic fungi, the requirement for invasive growth is even more acute. In this Primer, we explore the mechanobiology of fungal invasive growth and the emerging view of the different mechanisms that fungal pathogens deploy to gain entry to host tissue. We focus mainly on plant pathogens, where recent experimental work has been most extensive, and highlight key research questions for the future.

|
Scooped by
The Sainsbury Lab
July 16, 5:36 AM
|
Plant nucleotide-binding, leucine-rich repeat (NLR) immune receptors recognize pathogen effectors and activate defense. NLR genes can be non-functiona…

|
Scooped by
The Sainsbury Lab
July 16, 5:08 AM
|
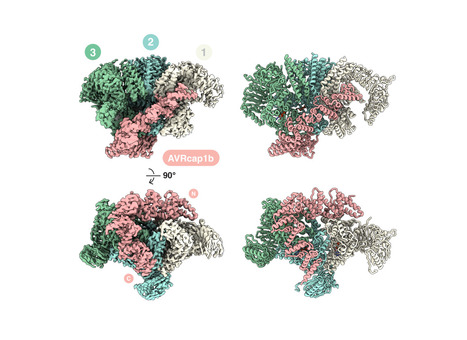
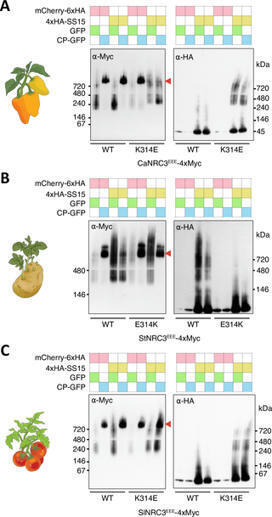
Helper NLRs function as central nodes in plant immune networks. Upon activation, they oligomerize into inflammasome like resistosomes to initiate immune signaling, yet the dynamics of resistosome assembly remain poorly understood. Here, we show that the virulence effector AVRcap1b from the Irish potato famine pathogen Phytophthora infestans suppresses immune activation by directly engaging oligomerization intermediates of the tomato helper NLR SlNRC3. CryoEM structures of SlNRC3 in AVRcap1b bound and unbound states reveal that AVRcap1b bridges multiple protomers, stabilizing a stalled intermediate and preventing formation of a functional resistosome. Leveraging AVRcap1b as a molecular tool, we also capture an additional SlNRC3 resistosome intermediate showing that assembly proceeds in a stepwise manner from dissociated monomers. These findings uncover a previously unrecognized vulnerability in NLR activation and reveal a pathogen strategy that disrupts immune complex assembly. This work advances mechanistic understanding of resistosome formation and uncovers a previously unrecognized facet of pathogen-plant coevolution.

|
Scooped by
The Sainsbury Lab
April 28, 3:41 PM
|
Key message Plant U-box E3 ligases PUB20 and PUB21 are flg22-triggered signaling components and negatively regulate immune responses. Abstract Plant U-box proteins (PUBs) constitute a class of E3 ligases that are associated with various stress responses. Among the class IV PUBs featuring C-terminal Armadillo (ARM) repeats, PUB20 and PUB21 are closely related homologs. Here, we show that both PUB20 and PUB21 negatively regulate innate immunity in plants. Loss of PUB20 and PUB21 function leads to enhanced resistance to surface inoculation with the virulent bacterium Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). However, the resistance levels remain unaffected after infiltration inoculation, suggesting that PUB20 and PUB21 primarily function during the early defense stages. The enhanced resistance to Pst DC3000 in PUB mutant plants (pub20-1, pub21-1, and pub20-1/pub21-1) correlates with extensive flg22-triggered reactive oxygen production, strong MPK3 activation, and enhanced transcriptional activation of early immune response genes. Additionally, PUB mutant plants (except pub21-1) exhibit constitutive stomatal closure after Pst DC3000 inoculation, implying the significant role of PUB20 in stomatal immunity. Comparative analyses of flg22 responses between PUB mutants and wild-type plants reveals that the robust activation of the pattern-induced immune responses may enhance resistance against Pst DC3000. Notably, the hypersensitivity responses triggered by RPM1/avrRpm1 and RPS2/avrRpt2 are independent of PUB20 and PUB21. These results suggest that PUB20 and PUB21 knockout mutations affect bacterial invasion, likely during the early stages, acting as negative regulators of plant immunity.

|
Scooped by
The Sainsbury Lab
April 28, 3:12 PM
|
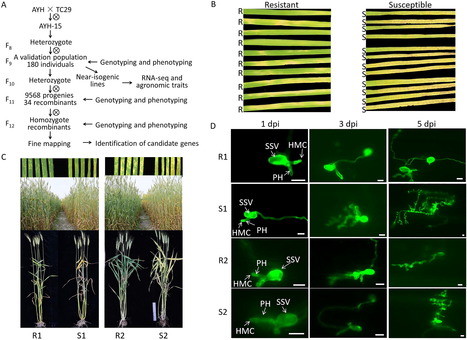
Stripe rust, caused by Puccinia striiformis f. sp. tritici ( Pst), is a devastating disease in wheat worldwide. Discovering and characterizing new resistance genes/QTL is crucial for wheat breeding programs. In this study, we fine-mapped and characterized a stripe rust resistance gene, YRAYH, on chromosome arm 5BL in the Chinese wheat landrace Anyuehong (AYH). Evaluations of stripe rust response to prevalent Chinese Pst races in near-isogenic lines derived from a cross of Anyuehong and Taichung 29 showed that YrAYH conferred a high level of resistance at all growth stages. Fine mapping using a large segregating population of 9748 plants, narrowed the YRAYH locus to a 3.7 Mb interval on chromosome arm 5BL that included 61 annotated genes. Transcriptome analysis of two NIL pairs identified 64 upregulated differentially expressed genes (DEGs) in the resistant NILs (NILs-R). Annotations indicated that many of these genes have roles in plant disease resistance pathways. Through a combined approach of fine-mapping and transcriptome sequencing, we identified a serine/threonine-protein kinase SRPK as a candidate gene underlying YrAYH. A unique 25 bp insertion was identified in the NILs-R compared to the NILs-S and previously published wheat genomes. An InDel marker was developed and co-segregated with YrAYH. Agronomic trait evaluation of the NILs suggested that YrAYH not only reduces the impact of stripe rust but was also associated with a gene that increases plant height and spike length.

|
Scooped by
The Sainsbury Lab
February 12, 5:01 AM
|
-
Asian soybean rust (ASR), caused by the obligate biotrophic fungus, Phakopsora pachyrhizi, was first reported in the continental United States of America (USA) in 2004 and over the years has been of concern to soybean production in the USA. The prevailing hypothesis is that P. pachyrhizi spores were introduced into the USA via hurricanes originating from South America, particularly Hurricane Ivan. -
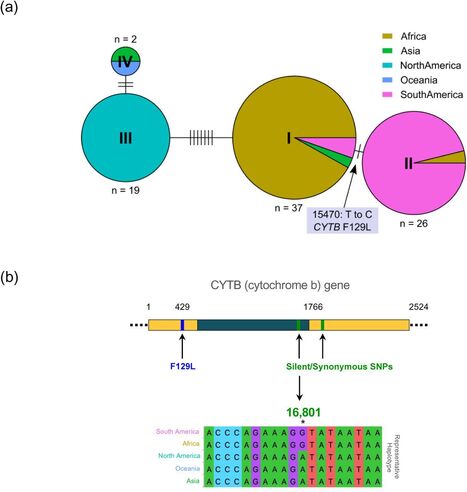
To investigate the genetic diversity and global population structure of P. pachyrhizi, we employed exome-capture based sequencing on 84 field isolates collected from different geographic regions worldwide. We compared the gene-encoding regions from all these field isolates and found that four major haplotypes are prevalent worldwide. Here, we provide genetic evidence supporting multiple incursions that have led to the currently established P. pachyrhizi population of the USA. Phylogenetic analysis of mitochondrial genes further supports this hypothesis. -
Notably, we observed limited genetic diversity in P. pachyrhizi populations in Brazil, suggesting a clonal population structure in that country that contrasts to populations from the USA and Africa. -
This study provides the first comprehensive characterization of P. pachyrhizi population structures defined by genetic evidence from populations across major soybean growing regions.

|
Scooped by
The Sainsbury Lab
February 12, 4:43 AM
|
Carbohydrate-based cell wall signaling impacts plant growth, development, and stress responses; however, how cell wall signals are perceived and transduced remains poorly understood. Several cell wall breakdown products have been described as typical damage-associated molecular patterns that activate plant immunity, including pectin-derived oligogalacturonides (OGs). Receptor kinases of the WALL-ASSOCIATED KINASE (WAK) family bind pectin and OGs and were previously proposed as OG receptors. However, unambiguous genetic evidence for the role of WAKs in OG responses is lacking. Here, we investigated the role of Arabidopsis (Arabidopsis thaliana) WAKs in OG perception using a clustered regularly interspaced short palindromic repeats mutant in which all 5 WAK genes were deleted. Using a combination of immune assays for early and late pattern-triggered immunity, we show that WAKs are dispensable for OG-induced signaling and immunity, indicating that they are not bona fide OG receptors.

|
Scooped by
The Sainsbury Lab
February 12, 4:40 AM
|
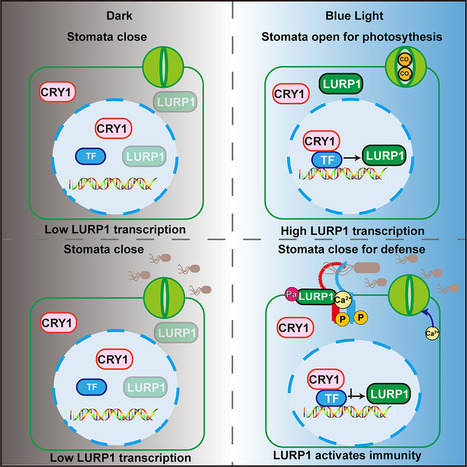
Plant stomata open in response to blue light, allowing gas exchange and water transpiration. However, open stomata are potential entry points for pathogens. Whether plants can sense pathogens and mount defense responses upon stomatal opening and how blue-light cues are integrated to balance growth-defense trade-offs are poorly characterized. We show that the Arabidopsis blue-light photoreceptor CRYPTOCHROME 1 (CRY1) mediates various aspects of immunity, including pathogen-triggered stomatal closure as well as activation of plant immunity through a typical light-responsive protein LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA (LURP1). LURP1 undergoes N-terminal palmitoylation in the presence of bacterial flagellin, prompting a change in subcellular localization from the cytoplasm to plasma membrane, where it enhances the activity of the receptor FLAGELLIN SENSING 2 (FLS2) to mediate plant defense. Collectively, these findings reveal that blue light regulates stomatal defense and highlight the dual functions of CRY1 in photosynthesis and immunity.

|
Scooped by
The Sainsbury Lab
February 12, 4:31 AM
|
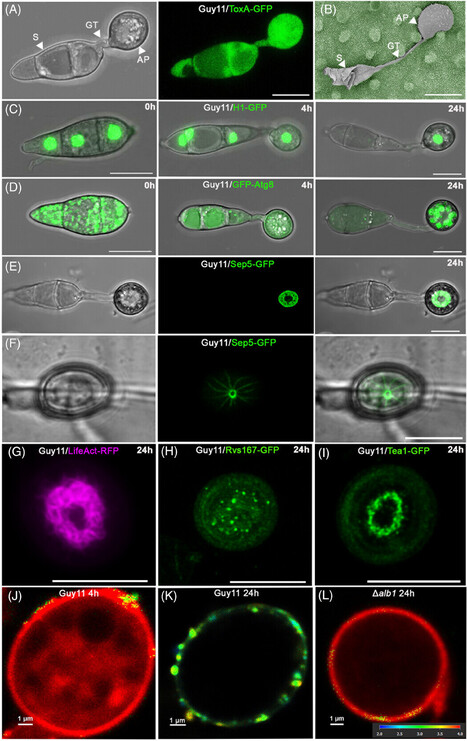
Magnaporthe oryzae is the causal agent of rice blast, one of the most serious diseases affecting rice cultivation around the world. During plant infection, M. oryzae forms a specialised infection structure called an appressorium. The appressorium forms in response to the hydrophobic leaf surface and relies on multiple signalling pathways, including a MAP kinase phosphorelay and cAMP-dependent signalling, integrated with cell cycle control and autophagic cell death of the conidium. Together, these pathways regulate appressorium morphogenesis.The appressorium generates enormous turgor, applied as mechanical force to breach the rice cuticle. Re-polarisation of the appressorium requires a turgor-dependent sensor kinase which senses when a critical threshold of turgor has been reached to initiate septin-dependent re-polarisation of the appressorium and plant infection. Invasive growth then requires differential expression and secretion of a large repertoire of effector proteins secreted by distinct secretory pathways depending on their destination, which is also governed by codon usage and tRNA thiolation. Cytoplasmic effectors require an unconventional Golgi-independent secretory pathway and evidence suggests that clathrin-mediated endocytosis is necessary for their delivery into plant cells. The blast fungus then develops a transpressorium, a specific invasion structure used to move from cell-to-cell using pit field sites containing plasmodesmata, to facilitate its spread in plant tissue. This is controlled by the same MAP kinase signalling pathway as appressorium development and requires septin-dependent hyphal constriction. Recent progress in understanding the mechanisms of rice infection by this devastating pathogen using live cell imaging procedures are presented.
|

|
Scooped by
The Sainsbury Lab
July 16, 5:56 AM
|
Author summary Plants have immune systems that protect them from various pathogens, including bacteria, fungi and nematodes. This immune system harnesses proteins called NLRs (nucleotide-binding and leucine-rich repeat proteins) to detect and respond to pathogens. However, pathogens often evolve strategies to suppress plant immunity. For example, the cyst nematode Globodera rostochiensis produces an effector protein called SPRYSEC15 (SS15) that inhibits certain NLR proteins, suppressing the plant’s immune response. In this study, we explored how a group of NLR proteins called NRCs, found in the Solanaceae plant family, have evolved to counteract SS15 inhibition. We discovered that while SS15 inhibits all tested NRC2 orthologs, some natural variants of NRC1 and NRC3 are insensitive to its effects. By reconstructing the evolutionary history of NRC3, we found that these proteins acquired insensitivity to SS15 through a specific amino acid substitution about 19 million years ago. Our findings highlight a previously understudied type of evolutionary arms race between plants and pathogens, where plants adapt to evade pathogen-derived suppressors. This research expands our understanding of plant-pathogen coevolution and offers insights that could guide the development of crops with improved disease resistance.

|
Scooped by
The Sainsbury Lab
July 16, 5:54 AM
|
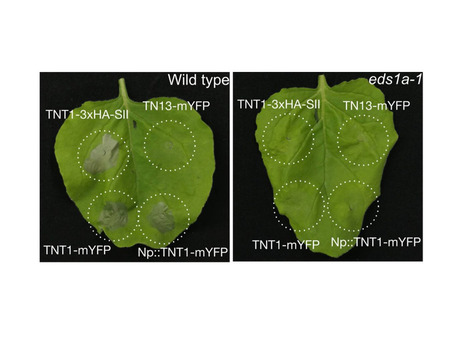
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors detect pathogen-secreted effectors inside host cells and induce a robust immune response, typically involving hypersensitive cell death. NLR genes are often genetically linked and can function as pairs or within larger NLR networks. We previously showed that the truncated Toll/Interleukin-1 Receptor (TIR)-type NLR TIR-NB13 (TN13) is required for resistance of Arabidopsis to Pseudomonas syringae pv. tomato (Pst) DC3000 lacking the type-III effector proteins AvrPto and AvrPtoB. TN13 is genetically linked to a full length TIR-NB-LRR (TNL) gene on chromosome 3. Here, we show that TN13, and its genetically linked TNL both localize to the ER membrane via N-terminal transmembrane domains, are required for resistance to Pst DC3000 (ΔAvrPto/AvrPtoB) and interact with each other in transient expression assays in Nicotiana benthamiana. In contrast to TN13, the full length TNL, which we named TN13-INTERACTING TNL1 (TNT1), induces an autoactive cell death response when expressed in N. benthamiana that depends on an atypical MHV motif in its NB-ARC domain, as well as the EDS1/SAG101/NRG1 module. TN13 and TNT1 furthermore interact with phylogenetically related NLRs encoded by a segmentally duplicated region on chromosome 5. Our data suggest that both TN13 and the genetically linked TNT1 could be part of a larger TIR-type NLR immune regulatory network, in which TNT1 contributes to basal immunity and might function as an autoactive death switch to induce cell death upon pathogen detection.

|
Scooped by
The Sainsbury Lab
July 16, 5:51 AM
|
Phakopsora pachyrhizi, an obligate biotrophic rust fungus, is the causal agent of Asian Soybean Rust (ASR) disease. Here, we utilized whole-genome dat…

|
Scooped by
The Sainsbury Lab
July 16, 5:46 AM
|
The receptor-like cytoplasmic kinase BIK1 and its close homolog PBL1 have been widely recognized as central components of plant immunity. However, most genetic studies of BIK1 and PBL1 functions were carried out with single T-DNA insertional mutant alleles. Some phenotypes observed in these mutants, e.g. autoimmunity, have been difficult to reconcile with the proposed role of BIK1 and PBL1 in pattern-triggered immunity. In this study, we generated multiple new alleles of bik1 and pbl1 by CRISPR-Cas9-based gene editing and systematically analyzed these mutants alongside existing T-DNA insertional lines. These analyses reinforced the central role of BIK1 and PBL1 in pattern-triggered immunity mediated by both receptor kinases and receptor-like proteins. At the same time, however, we revealed several pleiotropic phenotypes associated with T-DNA insertions that are not necessarily linked to loss of BIK1 or PBL1 function. Further analyses of newly generated bik1 pbl1 double mutants uncovered an even greater contribution of these kinases to immune signaling and disease resistance than previously appreciated. These findings clarify longstanding ambiguities surrounding BIK1 and PBL1 functions.

|
Scooped by
The Sainsbury Lab
July 16, 5:41 AM
|
Plant nucleotide-binding, leucine-rich-repeat (NLR) immune receptors recognize pathogen effectors and activate immunity. The NLR RPS2 recognizes AvrRpt2, a Pseudomonas effector that promotes virulence by proteolytically cleaving a membrane-tethered host protein, RIN4. RIN4 cleavage by AvrRpt2 generates fragments that activate RPS2. A model for RPS2 activation by RIN4 destruction is consistent with the ectopic activity of RPS2 in plants lacking RIN4 but does not explain the link between AvrRpt2’s virulence activity and RPS2 activation. We found that non-membrane-tethered RIN4 derivatives are potent cytosolic activators of RPS2. Activation of RPS2 by these RIN4 derivatives, like AvrRpt2-induced activation, and unlike ectopic activation in the absence of RIN4, requires the defense signaling protein NDR1. Cleavage products of RIN4 produced by AvrRpt2 play contrasting roles in the activation of RPS2, with the membrane-tethered C-terminal fragment suppressing RPS2 and the non-membrane-tethered internal fragment, dependent on compatibility with the C-terminal fragment, overcoming its suppression of RPS2.

|
Scooped by
The Sainsbury Lab
July 16, 5:32 AM
|
Introducing and characterizing variation through mutagenesis plus functional genomics can accelerate resistance breeding as well as our understanding of crop plant immunity. To reveal new germplasm resources for fungal disease resistance breeding in elite durum wheat, we challenged the diverse alleles in a sequenced and cataloged ethyl methanesulfonate mutagenized population of elite tetraploid wheat Triticum turgidum subsp. durum cv ‘Kronos’ with stripe rust. We screened 2,000 mutant lines and identified sixteen enhanced disease resistance (EDR) lines with persistent resistance to stripe rust over four years of field testing. To find broad-spectrum resistance, we challenged these lines with other major biotrophic and necrotrophic pathogens, including those causing Septoria tritici blotch, tan spot, Fusarium head blight and leaf rust. Enhanced resistance to multiple fungi was found in 13 of 16 EDR lines. Five EDR lines showed spontaneous lesion formation in the absence of pathogens, providing new mutant resources to study plant stress response in the absence of the confounding effects of pathogen infection. We mapped exome capture sequencing data of the EDR lines to a recently released long-read Kronos genome to aid in the identification of causal mutations. We located an EDR resistance locus to an 175 Mb interval on chromosome 1B. Importantly, these phenotypically characterized EDR lines are newly described durum germplasm coupled with improved functional genomics resources that are readily available for both wheat fungal resistance breeding and basic plant immunity research.

|
Scooped by
The Sainsbury Lab
July 7, 11:43 AM
|
Plant nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) confer disease resistance to many foliar and root parasites. However, the extent to which NLR-mediated immunity is differentially regulated between plant organs is poorly known. Here, we show that a large cluster of tomato (Solanum lycopersicum) genes, encoding the cyst and root-knot nematode disease resistance proteins Hero and MeR1 as well as the NLR helper NLR required for cell death 6 (NRC6), is nearly exclusively expressed in the roots. This root-specific gene cluster emerged in Solanum species about 21 million years ago through gene duplication of the ancient asterid NRC network. NLR sensors in this gene cluster function exclusively through NRC6 helpers to trigger hypersensitive cell death. These findings indicate that the NRC6 gene cluster has sub-functionalized from the larger NRC network to specialize in mediating resistance against root pathogens, including cyst and root-knot nematodes. We propose that some NLR gene clusters and networks may have evolved organ-specific gene expression as an adaptation to particular parasites and to reduce the risk of autoimmunity.

|
Scooped by
The Sainsbury Lab
April 28, 3:26 PM
|
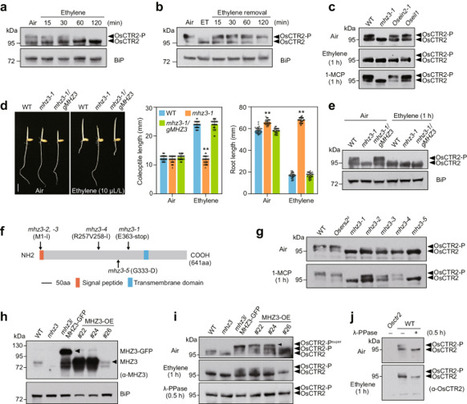
Ethylene regulates plant growth, development, and stress adaptation. However, the early signaling events following ethylene perception, particularly in the regulation of ethylene receptor/CTRs (CONSTITUTIVE TRIPLE RESPONSE) complex, remains less understood. Here, utilizing the rapid phospho-shift of rice OsCTR2 in response to ethylene as a sensitive readout for signal activation, we revealed that MHZ3, previously identified as a stabilizer of ETHYLENE INSENSITIVE 2 (OsEIN2), is crucial for maintaining OsCTR2 phosphorylation. Genetically, both functional MHZ3 and ethylene receptors prove essential for OsCTR2 phosphorylation. MHZ3 physically interacts with both subfamily I and II ethylene receptors, e.g., OsERS2 and OsETR2 respectively, stabilizing their association with OsCTR2 and thereby maintaining OsCTR2 activity. Ethylene treatment disrupts the interactions within the protein complex MHZ3/receptors/OsCTR2, reducing OsCTR2 phosphorylation and initiating downstream signaling. Our study unveils the dual role of MHZ3 in fine-tuning ethylene signaling activation, providing insights into the initial stages of the ethylene signaling cascade. The early signalling events following ethylene perception by plants remain incompletely understood. Here the authors show that in the absence of ethylene, rice MHZ3, a known stabilizer of OsEIN2, promotes phosphorylation of OsCTR2 to suppress ethylene signalling.

|
Scooped by
The Sainsbury Lab
February 12, 5:09 AM
|
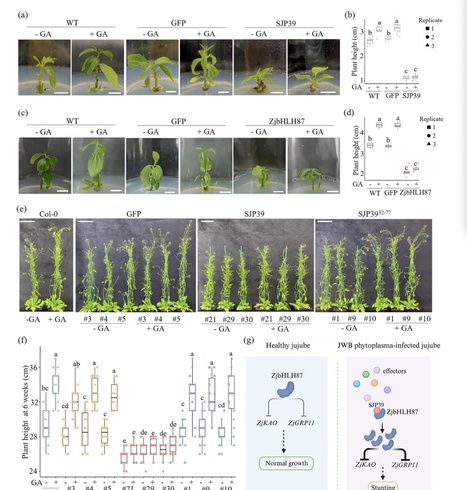
-
Phytoplasmas are specialized phloem-limited bacteria that cause diseases on various crops resulting in significant agricultural losses. This research focuses on the jujube Witches’ Broom (JWB) phytoplasma and investigates the host-manipulating activity of the effector SJP39. -
We found that SJP39 directly interacts with the plant transcription factor bHLH87 in the nuclei. SJP39 stabilizes the bHLH87 homologs in A. thaliana and jujube, leading to growth defects in the plants. -
Transcriptomic analysis indicates that SJP39 affects the gibberellin (GA) pathway in jujube. We further demonstrate that ZjbHLH87 regulates GA signalling as a negative regulator and SJP39 enhances this regulation. -
The research offers important insights into the pathogenesis of JWB disease and identified SJP39 as a virulence factor that can contribute to the growth defects caused by JWB phytoplasma infection. These findings open new opportunities to manage JWB and other phytoplasma diseases.

|
Scooped by
The Sainsbury Lab
February 12, 4:48 AM
|
The ability of plants to perceive and react to biotic and abiotic stresses is critical for their health. We recently identified a core set of genes consistently induced by members of the leaf microbiota, termed general non-self response (GNSR) genes. Here we show that GNSR components conversely impact leaf microbiota composition. Specific strains that benefited from this altered assembly triggered strong plant responses, suggesting that the GNSR is a dynamic system that modulates colonization by certain strains. Examination of the GNSR to live and inactivated bacteria revealed that bacterial abundance, cellular composition and exposure time collectively determine the extent of the host response. We link the GNSR to pattern-triggered immunity, as diverse microbe- or danger-associated molecular patterns cause dynamic GNSR gene expression. Our findings suggest that the GNSR is the result of a dose-responsive perception and signalling system that feeds back to the leaf microbiota and contributes to the intricate balance of plant–microbiome interactions. The plant general non-self response system is triggered by leaf microbiota members and, in turn, impacts their colonization.

|
Scooped by
The Sainsbury Lab
February 12, 4:41 AM
|
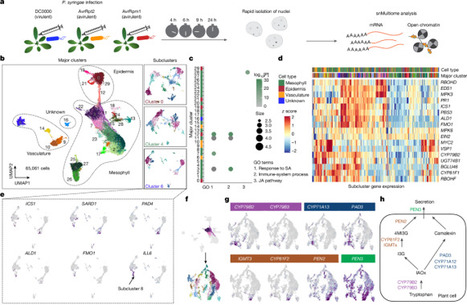
Plants lack specialized and mobile immune cells. Consequently, any cell type that encounters pathogens must mount immune responses and communicate with surrounding cells for successful defence. However, the diversity, spatial organization and function of cellular immune states in pathogen-infected plants are poorly understood1. Here we infect Arabidopsis thaliana leaves with bacterial pathogens that trigger or supress immune responses and integrate time-resolved single-cell transcriptomic, epigenomic and spatial transcriptomic data to identify cell states. We describe cell-state-specific gene-regulatory logic that involves transcription factors, putative cis-regulatory elements and target genes associated with disease and immunity. We show that a rare cell population emerges at the nexus of immune-active hotspots, which we designate as primary immune responder (PRIMER) cells. PRIMER cells have non-canonical immune signatures, exemplified by the expression and genome accessibility of a previously uncharacterized transcription factor, GT-3A, which contributes to plant immunity against bacterial pathogens. PRIMER cells are surrounded by another cell state (bystander) that activates genes for long-distance cell-to-cell immune signalling. Together, our findings suggest that interactions between these cell states propagate immune responses across the leaf. Our molecularly defined single-cell spatiotemporal atlas provides functional and regulatory insights into immune cell states in plants. The development of a molecularly defined spatiotemporal atlas of pathogen-infected Arabidopsis thaliana leaves reveals specific cell states that have distinct roles in plant immunity.

|
Scooped by
The Sainsbury Lab
February 12, 4:36 AM
|
Fungi are the most important group of plant pathogens, responsible for many of the world’s most devastating crop diseases. One of the reasons they are such successful pathogens is because several fungi have evolved the capacity to breach the tough outer cuticle of plants using specialized infection structures called appressoria. This is exemplified by the filamentous ascomycete fungus Magnaporthe oryzae, causal agent of rice blast, one of the most serious diseases affecting rice cultivation globally. M. oryzae develops a pressurized dome-shaped appressorium that uses mechanical force to rupture the rice leaf cuticle. Appressoria form in response to the hydrophobic leaf surface, which requires the Pmk1 MAP kinase signalling pathway, coupled to a series of cell-cycle checkpoints that are necessary for regulated cell death of the fungal conidium and development of a functionally competent appressorium. Conidial cell death requires autophagy, which occurs within each cell of the spore, and is regulated by components of the cargo-independent autophagy pathway. This results in trafficking of the contents of all three cells to the incipient appressorium, which develops enormous turgor of up to 8.0 MPa, due to glycerol accumulation, and differentiates a thickened, melanin-lined cell wall. The appressorium then re-polarizes, re-orienting the actin and microtubule cytoskeleton to enable development of a penetration peg in a perpendicular orientation, that ruptures the leaf surface using mechanical force. Re-polarization requires septin GTPases which form a ring structure at the base of the appressorium, which delineates the point of plant infection, and acts as a scaffold for actin re-localization, enhances cortical rigidity, and forms a lateral diffusion barrier to focus polarity determinants that regulate penetration peg formation. Here we review the mechanism of regulated cell death in M. oryzae, which requires autophagy but may also involve ferroptosis. We critically evaluate the role of regulated cell death in appressorium morphogenesis and examine how it is initiated and regulated, both temporally and spatially, during plant infection. We then use this synopsis to present a testable model for control of regulated cell death during appressorium-dependent plant infection by the blast fungus.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...