Your new post is loading...
 Your new post is loading...

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:14 PM
|
This post is adapted from a talk I gave at the Plantae Presents webinar More Money, More Data: Data-Driven versus Hypothesis-Driven Research (recording here). The webinar was moderated by Plantae Fellows Krishna Alamuru, Dennis Baffour-Awuah, Aditi Bhat, and Kestrel Maio. They moderated the discussion and invited me and Ivan Baxter of the Donald Danforth Plant Science Center to explore the evolving landscape of plant biology in the era of big data. My contribution tackled the presumed tension between traditional hypothesis-driven science and big, data-driven approaches. The slides from my presentation are available on Zenodo. What follows is a blog version of that talk.

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:12 PM
|
A mentoring event at a scientific Congress called for better support of early-career scientists — but what followed sent mixed signals. This blog reflects on those moments and ends with a pledge: stop displaying journal names on your slides.

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:10 PM
|
What would you do if war and famine broke out in your homeland — while you were ten thousand kilometers away, working on your PhD? This is what Hagos Mohammedseid Juhar faced. And this is how he chose to act.

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:08 PM
|
In the harsh deserts of southern Algeria, scientists like Nadjette Djemouai are uncovering the power of native microbes to support sustainable agriculture. With support from GetGenome, they’re bringing genomics to the frontlines of climate resilience.
Nucleotide binding and leucine-rich repeat (NLR) proteins are intracellular immune receptors that occur across all kingdoms of life but are particularly highly diversified in plants (Barragan & Weigel, 2021). In plants, NLRs that carry a coiled-coil (CC) domain at their N termini are the most phylogenetically widespread class. Following pathogen recognition, CC-NLR proteins oligomerize into pentameric or hexameric pore-like complexes (Wang et al., 2019; Förderer et al., 2022; Zhao et al., 2022; Liu et al., 2024; Madhuprakash et al., 2024). These complexes, known as resistosomes, are a defining feature of NLRs that execute the immune response; some of them are known to translocate to cellular membranes and trigger immune responses such as calcium influx and hypersensitive cell death (Duggan et al., 2021; Contreras et al., 2022; Ibrahim et al., 2024). The prevailing model is that the funnel-shaped structure of CC-NLR resistosomes inserts into membranes and is required for executing the cell death and immune response (Wang et al., 2019; Adachi et al., 2019b; Förderer & Kourelis, 2023). This funnel-shaped structure is formed by the N-terminal α1 helix, which is a structurally dynamic region that is difficult to resolve using cryo-electron microscopy (cryo-EM) (Förderer et al., 2022; Zhao et al., 2022; Liu et al., 2024; Madhuprakash et al., 2024).
Via The Sainsbury Lab
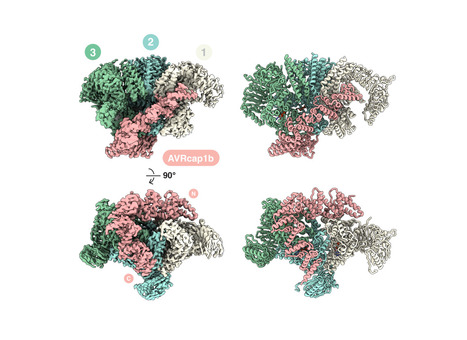
Helper NLRs function as central nodes in plant immune networks. Upon activation, they oligomerize into inflammasome like resistosomes to initiate immune signaling, yet the dynamics of resistosome assembly remain poorly understood. Here, we show that the virulence effector AVRcap1b from the Irish potato famine pathogen Phytophthora infestans suppresses immune activation by directly engaging oligomerization intermediates of the tomato helper NLR SlNRC3. CryoEM structures of SlNRC3 in AVRcap1b bound and unbound states reveal that AVRcap1b bridges multiple protomers, stabilizing a stalled intermediate and preventing formation of a functional resistosome. Leveraging AVRcap1b as a molecular tool, we also capture an additional SlNRC3 resistosome intermediate showing that assembly proceeds in a stepwise manner from dissociated monomers. These findings uncover a previously unrecognized vulnerability in NLR activation and reveal a pathogen strategy that disrupts immune complex assembly. This work advances mechanistic understanding of resistosome formation and uncovers a previously unrecognized facet of pathogen-plant coevolution.
Via The Sainsbury Lab
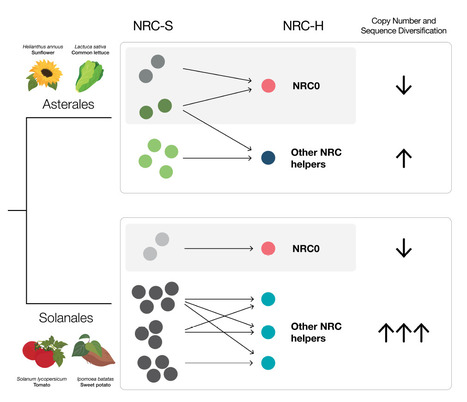
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) are known for their rapid evolution, even at the intraspecific level, yet the rates of evolution differ significantly across various NLR classes. Within the NRC (NLR Required for Cell Death) network, NLRs operate in complex sensor-helper configurations to confer immunity against a diverse array of pathogens, particularly in Asterids. While helper NLRs are typically conserved and evolve slowly, sensor NLRs tend to evolve more rapidly. However, the functional connections between slow and fast-evolving NLRs remain poorly understood, notably in important crop species. We conducted a comparative analysis of NLRs across 40 Solanales and 29 Asterales genomes to explore NRC network expansion and diversification within the less-studied Asterales order. Our findings reveal that the NRC network has expanded less in Asterales compared to Solanales. We functionally validated a minimal Asterales NRC network with 2 helpers and 9 sensors in common lettuce (Lactuca sativa). Through selection and diversification analysis and structural modeling of NRC helper and sensor subclades in the Lactuca genus, we found varying evolutionary diversification rates between NRC helpers and sensors. We found a correlation between sensor diversification rates and helper dependency, with sensors reliant on a phylogenetically conserved helpers experiencing limited diversification pressure. Our results highlight the lineage- and function-specific evolution of the NRC network, offering insights into the evolutionary pressures shaping plant immune receptor networks.
Via The Sainsbury Lab
Author summary Plants have immune systems that protect them from various pathogens, including bacteria, fungi and nematodes. This immune system harnesses proteins called NLRs (nucleotide-binding and leucine-rich repeat proteins) to detect and respond to pathogens. However, pathogens often evolve strategies to suppress plant immunity. For example, the cyst nematode Globodera rostochiensis produces an effector protein called SPRYSEC15 (SS15) that inhibits certain NLR proteins, suppressing the plant’s immune response. In this study, we explored how a group of NLR proteins called NRCs, found in the Solanaceae plant family, have evolved to counteract SS15 inhibition. We discovered that while SS15 inhibits all tested NRC2 orthologs, some natural variants of NRC1 and NRC3 are insensitive to its effects. By reconstructing the evolutionary history of NRC3, we found that these proteins acquired insensitivity to SS15 through a specific amino acid substitution about 19 million years ago. Our findings highlight a previously understudied type of evolutionary arms race between plants and pathogens, where plants adapt to evade pathogen-derived suppressors. This research expands our understanding of plant-pathogen coevolution and offers insights that could guide the development of crops with improved disease resistance.
Via The Sainsbury Lab
Helper NLRs function as central nodes in plant immune networks. Upon activation, they oligomerize into inflammasome like resistosomes to initiate immune signaling, yet the dynamics of resistosome assembly remain poorly understood. Here, we show that the virulence effector AVRcap1b from the Irish potato famine pathogen Phytophthora infestans suppresses immune activation by directly engaging oligomerization intermediates of the tomato helper NLR SlNRC3. CryoEM structures of SlNRC3 in AVRcap1b bound and unbound states reveal that AVRcap1b bridges multiple protomers, stabilizing a stalled intermediate and preventing formation of a functional resistosome. Leveraging AVRcap1b as a molecular tool, we also capture an additional SlNRC3 resistosome intermediate showing that assembly proceeds in a stepwise manner from dissociated monomers. These findings uncover a previously unrecognized vulnerability in NLR activation and reveal a pathogen strategy that disrupts immune complex assembly. This work advances mechanistic understanding of resistosome formation and uncovers a previously unrecognized facet of pathogen-plant coevolution.
Via The Sainsbury Lab
As much as 10% of plant immune receptors from the nucleotide-binding domain leucine-rich repeat (NLR) family carry integrated domains (IDs) that can directly bind pathogen effectors. However, it remains unclear whether direct binding to effectors is a universal feature of ID-containing NLRs given that only a few NLR-IDs have been functionally characterized. Here we show that the rice (Oryza sativa) sensor NLR-ID Pii2 confers resistance to strains of the rice blast fungus Magnaporthe oryzae that carry the effector AVR-Pii without directly binding this protein. First, we show that AVR-Pii binds the exocyst subunit OsExo70F2 in rice (Oryza sativa) to dissociate preformed complexes of OsExo70F2 with host RPM1 INTERACTING PROTEIN4 (RIN4) at the conserved NOI motif, facilitating a possible virulence function. Second, we show that in its resting state, Pii2 binds OsExo70F2 and OsExo70F3, essential components of Pii-mediated resistance, through its integrated NOI domain. Remarkably, AVR-Pii binding to OsExo70F2/F3 leads to dissociation of the Pii2–OsExo70F2 and Pii2–OsExo70F3 complexes, destabilization of Pii2, and activation of immunity. These findings support a novel conceptual model in which an NLR-ID monitors alterations of tethered host proteins targeted by pathogen effectors, providing insight into pathogen recognition mechanisms.
Via The Sainsbury Lab
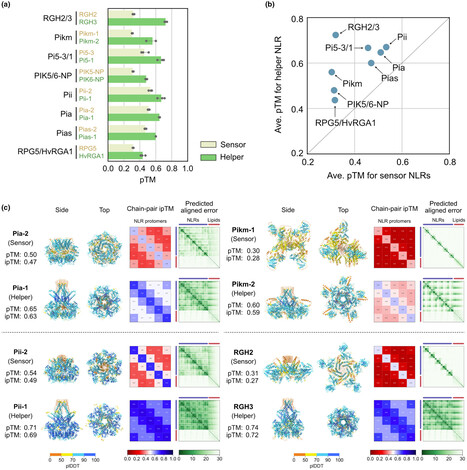
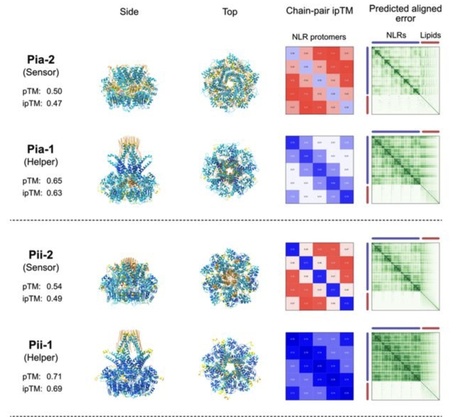
NLR immune receptors can be functionally organized in genetically linked sensor-helper pairs. However, methods to categorize paired NLRs remain limited, primarily relying on the presence of non-canonical domains in some sensor NLRs. Here, we propose that the AI system AlphaFold 3 can classify paired NLR proteins into sensor or helper categories based on predicted structural characteristics. Helper NLRs showed higher AlphaFold 3 confidence scores than sensors when modelled in oligomeric configurations. Furthermore, funnel-shaped structures—essential for activating immune responses—were reliably predicted in helpers but not in sensors. Applying this method to uncharacterized NLR pairs from rice, we found that AlphaFold 3 can differentiate between putative sensors and helpers even when both proteins lack non-canonical domain annotations. These findings suggest that AlphaFold 3 offers a new approach to categorize NLRs and enhances our understanding of the functional configurations in plant immune systems, even in the absence of non-canonical domain annotations.
Via The Sainsbury Lab
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins can engage in complex interactions to detect pathogens and execute a robust immune response via downstream helper NLRs. However, the biochemical mechanisms of helper NLR activation by upstream sensor NLRs remain poorly understood. Here, we show that the coiled-coil helper NLR NRC2 from Nicotiana benthamiana accumulates in vivo as a homodimer that converts into a higher-order oligomer upon activation by its upstream virus disease resistance protein Rx. The cryo-EM structure of NbNRC2 in its resting state revealed intermolecular interactions that mediate homodimer formation and contribute to immune receptor autoinhibition. These dimerization interfaces have diverged between paralogous NRC proteins to insulate critical network nodes and enable redundant immune pathways, possibly to minimise undesired cross-activation and evade pathogen suppression of immunity. Our results expand the molecular mechanisms of NLR activation pointing to transition from homodimers to higher-order oligomeric resistosomes.
Via The Sainsbury Lab
Abstract Pathogens have evolved sophisticated mechanisms to manipulate host cell membrane dynamics, a crucial adaptation to survive in hostile environments shaped by innate immune responses. Plant-derived membrane interfaces, engulfing invasive hyphal projections of fungal and oomycete pathogens, are prominent junctures dictating infection outcomes. Understanding how pathogens transform these host-pathogen interfaces to their advantage remains a key biological question. Here, we identified a conserved effector, secreted by plant pathogenic oomycetes, that co-opts a host Rab GTPase-activating protein (RabGAP), TOPGAP, to remodel the host-pathogen interface. The effector, PiE354, hijacks TOPGAP as a susceptibility factor to usurp its GAP activity on Rab8a, a key Rab GTPase crucial for defense-related secretion. By hijacking TOPGAP, PiE354 purges Rab8a from the plasma membrane, diverting Rab8a-mediated immune trafficking away from the pathogen interface. This mechanism signifies an uncanny evolutionary adaptation of a pathogen effector in co-opting a host regulatory component to subvert defense-related secretion, thereby providing unprecedented mechanistic insights into the reprogramming of host membrane dynamics by pathogens.
Via The Sainsbury Lab
|

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:13 PM
|
There are many toxic behaviors in academia, but one we rarely talk about is #AcademicPoaching. Have you ever been a victim?

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:11 PM
|
Publishing your poster creates a verifiable and citable record of your work. It also protects you from bad actors — the evidence will be there for all to see.

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:09 PM
|
In an age of growing tension between north and south, one ordinary orange butterfly flutters freely across continents, unbothered by border checkpoints. This is the story of the painted lady, Vanessa cardui.

|
Scooped by
Kamoun Lab @ TSL
August 9, 2:07 PM
|
You don’t need money or a title to influence others. We all hold a kind of soft power — just by how we choose to act.
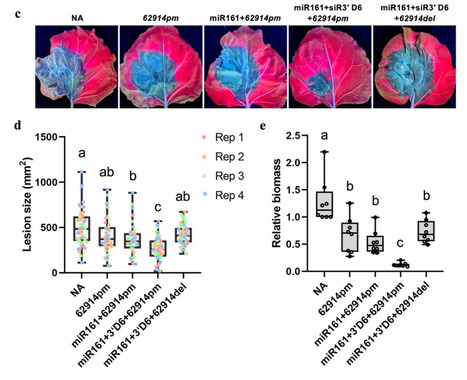
Small RNA-mediated gene silencing contributes to plant immunity. The secondary small interfering RNA (siRNA) pathway promotes defense by silencing target genes in invading fungal and oomycete pathogens. Many secondary siRNAs derive from transcripts potentially encoding pentatricopeptide repeat (PPR) proteins. Here, we report that siRNA production is an ancient function of an evolutionarily conserved clade of PPR genes that undergo extensive within-species diversification. In Arabidopsis thaliana, siRNA-source PPRs are physically clustered in one locus on Chromosome 1. These sequences are diversified through gene duplication followed by sequence diversification as well as accumulation of high-impact variations including pseudogenization. This diversity leads to the accumulation of a diverse PPR-siRNA pool, consistent with an engagement in a co-evolutionary arms race with the pathogens. This study defines siRNA-producing PPRs as a family of defense genes and highlights the potential of PPR-siRNA-based engineering for enhancing broad-spectrum disease resistance.
Via The Sainsbury Lab
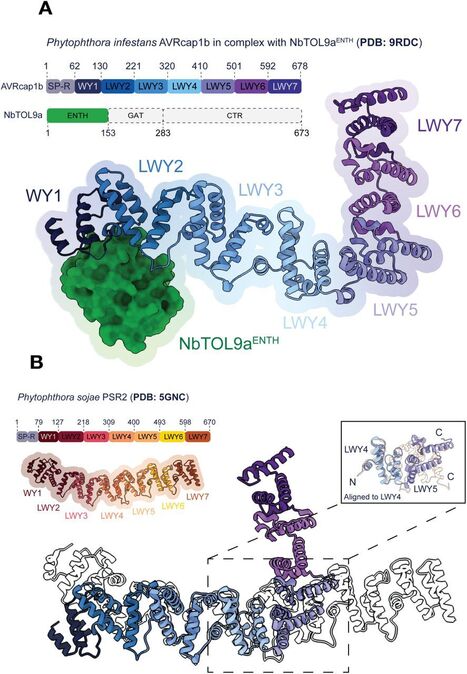
Pathogens counteract central nodes of NLR immune receptor networks to suppress immunity. However, the mechanisms by which pathogens hijack helper NLR pathways are poorly understood. Here, we show that an effector from the potato late blight pathogen Phytophthora infestans bridges the host protein NbTOL9a, a putative member of the host ESCRT pathway, to a helper NLR to suppress immunity. In this work, we solved the crystal structure of the RXLR-LWY effector AVRcap1b in complex with the ENTH domain of NbTOL9a. The structure revealed that unlike other RXLR-LWY effectors, AVRcap1b has a novel L-shaped fold that defines a new structural family of effectors in the Phytophthora genus. Moreover, we defined the AVRcap1b/NbTOL9a binding interface and designed effector mutants that don’t bind NbTOL9a, impairing immune suppression. This indicates that ENTH binding is required for full virulence activity of this effector. Lastly, we show that AVRcap1b associates specifically with activated NbNRC2 independently of NbTOL9a binding. This suggests that the effector functions as a bridge that interconnects NbNRC2 with the NbTOL9a pathway. These results illustrate an unprecedented pathogen mechanism to hijack helper NLR pathways and suppress immunity.
Via The Sainsbury Lab
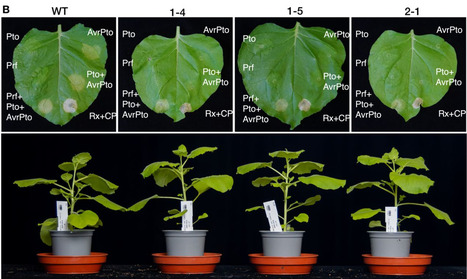
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) can function in networks of sensors and helpers to induce hypersensitive cell death and immunity against pathogens. The tomato sensor NLR Prf guards the Pto kinase from AvrPto and AvrPtoB effector perturbation and activates the downstream helpers NRC2 and NRC3. Prf is conserved across the Solanaceae and its ortholog in the model species Nicotiana benthamiana is also required for detection of AvrPto/AvrPtoB function on Pto. A recent study reported that cell death induction after transient expression of an autoactive mutant of tomato NRC3 is abolished upon RNAi silencing of Prf in N. benthamiana. Here we generated loss-of-function prf mutants in N. benthamiana and demonstrate that autoactive mutants of eight canonical tomato NRCs (NRC0, NRC1, NRC2, NRC3, NRC4a, NRC4b, NRC6, and NRC7) still induce hypersensitive cell death when expressed transiently in the prf mutant background. Autoactive tomato NRCs also triggered cell death when expressed in lettuce (Lactuca sativa), an Asteraceae plant that does not have a Prf ortholog. These results confirm a unidirectional dependency of sensors and helpers in the NRC network and underscore the value of the N. benthamiana and lettuce model systems for studying functional relationships between paired and networked NLRs.
Via The Sainsbury Lab
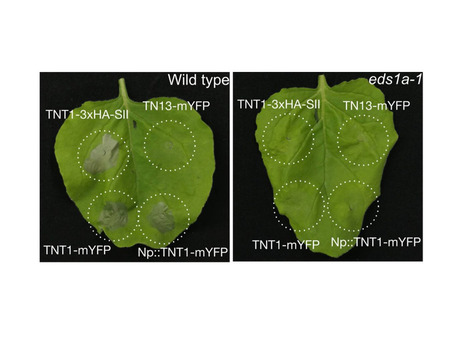
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors detect pathogen-secreted effectors inside host cells and induce a robust immune response, typically involving hypersensitive cell death. NLR genes are often genetically linked and can function as pairs or within larger NLR networks. We previously showed that the truncated Toll/Interleukin-1 Receptor (TIR)-type NLR TIR-NB13 (TN13) is required for resistance of Arabidopsis to Pseudomonas syringae pv. tomato (Pst) DC3000 lacking the type-III effector proteins AvrPto and AvrPtoB. TN13 is genetically linked to a full length TIR-NB-LRR (TNL) gene on chromosome 3. Here, we show that TN13, and its genetically linked TNL both localize to the ER membrane via N-terminal transmembrane domains, are required for resistance to Pst DC3000 (ΔAvrPto/AvrPtoB) and interact with each other in transient expression assays in Nicotiana benthamiana. In contrast to TN13, the full length TNL, which we named TN13-INTERACTING TNL1 (TNT1), induces an autoactive cell death response when expressed in N. benthamiana that depends on an atypical MHV motif in its NB-ARC domain, as well as the EDS1/SAG101/NRG1 module. TN13 and TNT1 furthermore interact with phylogenetically related NLRs encoded by a segmentally duplicated region on chromosome 5. Our data suggest that both TN13 and the genetically linked TNT1 could be part of a larger TIR-type NLR immune regulatory network, in which TNT1 contributes to basal immunity and might function as an autoactive death switch to induce cell death upon pathogen detection.
Via The Sainsbury Lab
Plant nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) confer disease resistance to many foliar and root parasites. However, the extent to which NLR-mediated immunity is differentially regulated between plant organs is poorly known. Here, we show that a large cluster of tomato (Solanum lycopersicum) genes, encoding the cyst and root-knot nematode disease resistance proteins Hero and MeR1 as well as the NLR helper NLR required for cell death 6 (NRC6), is nearly exclusively expressed in the roots. This root-specific gene cluster emerged in Solanum species about 21 million years ago through gene duplication of the ancient asterid NRC network. NLR sensors in this gene cluster function exclusively through NRC6 helpers to trigger hypersensitive cell death. These findings indicate that the NRC6 gene cluster has sub-functionalized from the larger NRC network to specialize in mediating resistance against root pathogens, including cyst and root-knot nematodes. We propose that some NLR gene clusters and networks may have evolved organ-specific gene expression as an adaptation to particular parasites and to reduce the risk of autoimmunity.
Via The Sainsbury Lab
Rice blast disease, caused by the fungus Magnaporthe oryzae, is one of the most devastating diseases of rice. Therefore, understanding the mechanisms of blast fungus infection and resistance of rice against the disease is important for global food security. In this study, we show that the M. oryzae effector protein AVR-PikD binds rice sHMA proteins and stabilizes them, presumably to enhance pathogen infection. We show that loss-of-function mutants in one rice sHMA, OsHIPP20, reduced the level of susceptibility against a compatible isolate of M. oryzae, suggesting that M. oryzae requires host sHMA to facilitate invasion. Remarkably, OsHIPP20 knockout rice line showed no growth defect, suggesting editing sHMA genes may present a novel source of resistance against blast disease.
Via The Sainsbury Lab
NRCs are essential helper NLR (nucleotide-binding domain and leucine-rich repeat) proteins that execute immune responses triggered by sensor NLRs. The resting state of NbNRC2 was recently shown to be a homodimer, but the sensor-activated state remains unclear. Using cryo-EM, we determined the structure of sensor-activated NbNRC2, which forms a hexameric inflammasome-like resistosome. Mutagenesis of the oligomerization interface abolished immune signaling, confirming the functional significance of the NbNRC2 resistosome. Comparative structural analyses between the resting state homodimer and sensor-activated homohexamer revealed substantial rearrangements, providing insights into NLR activation mechanisms. Furthermore, structural comparisons between NbNRC2 hexamer and previously reported CC-NLR pentameric assemblies revealed features allowing an additional protomer integration. Using the NbNRC2 hexamer structure, we assessed the recently released AlphaFold 3 for predicting activated CC-NLR oligomers, revealing high-confidence modeling of NbNRC2 and other CC-NLR amino-terminal α1 helices, a region proven difficult to resolve structurally. Overall, our work sheds light on NLR activation mechanisms and expands understanding of NLR structural diversity.
Via The Sainsbury Lab
Plants utilize complex immune systems to fend off invading pathogens. The nucleotide-binding domain and leucine-rich repeat (NLR) proteins function as intracellular immune receptors and play major roles in plant immunity. In solanaceous plants, several sensor NLRs form a complex genetic network with helper NLRs called NRCs (NLR-required for cell death) to trigger immune responses. However, the evolution of genetic compatibility between sensor NLRs and NRCs was unclear. Here, we showed that NRC3, one of the NRC subgroups in solanaceous plants, underwent subfunctionalization after the Solanum-Nicotiana divergence, altering the NRC network in Nicotiana. Using natural, chimeric, and reconstructed ancestral NRC variants, we mapped six critical residues on multiple protein surfaces of NRC3 contributing to subfunctionalization. These findings reveal how mutations in NRC alleles lead to subfunctionalization, altering sensor-helper NLR compatibility and increasing the complexity of plant immune systems.
Via The Sainsbury Lab
|



 Your new post is loading...
Your new post is loading...