Your new post is loading...
 Your new post is loading...

|
Scooped by
The Sainsbury Lab
August 18, 2025 7:50 AM
|
Asian soybean rust (ASR), caused by the obligate biotrophic fungus Phakopsora pachyrhizi, was first reported in the continental United States of America (USA) in 2004 and over the years has been of concern to soybean production in the United States. The prevailing hypothesis is that P. pachyrhizi spores were introduced into the United States via hurricanes originating from South America, particularly hurricane Ivan. To investigate the genetic diversity and global population structure of P. pachyrhizi, we employed exome-capture based sequencing on 84 field isolates collected from different geographic regions worldwide. We compared the gene-encoding regions from all these field isolates and found that four major mitochondrial haplotypes are prevalent worldwide. Here, we provide genetic evidence supporting multiple incursions that have led to the currently established P. pachyrhizi population of the United States. Phylogenetic analysis of mitochondrial genes further supports this hypothesis. We observed limited genetic diversity in P. pachyrhizi populations across different geographic regions, suggesting a clonal population structure. Additionally, this study is the first to report the F129L mutation in the Cytb gene outside South America, which is associated with strobilurin tolerance. This study provides the first comprehensive characterisation of P. pachyrhizi population structures defined by genetic evidence from populations across major soybean-growing regions.

|
Scooped by
The Sainsbury Lab
August 6, 2025 9:25 AM
|
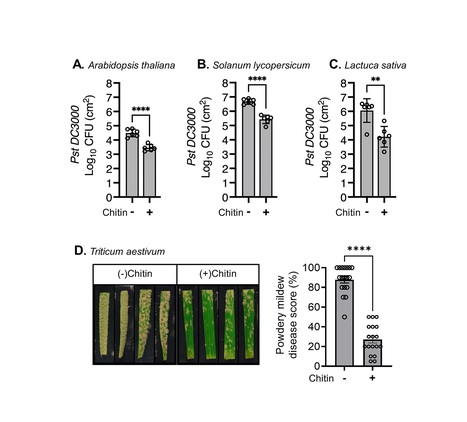
Chitin triggers localised and systemic plant immune responses, making it a promising treatment for sustainable disease resistance. However, the precise molecular mechanisms underlying chitin-induced systemic effects in plants remain unknown. In this study, we investigated the effects of soil amendment with crab chitin flakes (hereafter chitin) on pattern-triggered immunity (PTI) and systemic disease resistance in various plant species. We found that soil amendment with chitin potentiates PTI and disease resistance against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 in lettuce, tomato and Arabidopsis as well as against the fungal pathogen Blumeria graminis f. sp. tritici (Bgt), which causes powdery mildew in wheat. Using micrografting in Arabidopsis, we demonstrated that this systemic effect is dependent on active chitin perception in the roots. We also showed that induced systemic resistance (ISR) and pattern-recognition receptors (PRRs)/coreceptors, but not systemic acquired resistance (SAR), are involved in the systemic effects triggered by chitin soil amendment. These systemic effects correlated with the transcriptional upregulation of key PTI regulators in distal leaves upon chitin soil amendment. Notably, chitin-triggered systemic immunity was independent of microbes present in soil or chitin flakes. Together, these findings contribute to a better understanding of chitin-triggered systemic immunity, from active chitin perception in roots to the potentiation of PTI in the leaves, ultimately priming plants to mount enhanced defence responses against pathogen attacks. Our study provides valuable insights into the molecular mechanisms of chitin soil amendment and resulting induced immunity and highlights its potential use for sustainable crop protection strategies.

|
Scooped by
The Sainsbury Lab
July 31, 2025 4:42 AM
|
In the ongoing plant–pathogen arms race, plants use pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs), while in successful pathogens, PAMPs can evolve to evade detection. Engineering PRRs to recognize evading PAMPs could potentially generate broad-spectrum and durable disease resistance. Here we reverse-engineered two natural variants of the PRR FLAGELLIN SENSING 2 (FLS2), VrFLS2XL and GmFLS2b, with extended recognition specificities towards evading flg22 variants. We identified minimal gain-of-function residues enabling blind FLS2s to recognize otherwise evading flg22 variants. We uncovered two strategies: (1) optimizing FLS2–flg22 interaction around flg22’s key evasion sites and (2) strengthening direct FLS2–BAK1 interaction to overcome weak agonistic and antagonistic flg22s, respectively. In addition, we leveraged polymorphisms that enhance recognition through unknown mechanisms to engineer a superior recognition capability. These findings offer basic design principles to engineer PRRs with broader recognition spectra, paving the way for PRR engineering to generate precisely gene-edited disease-resistant crops. By dissecting natural variants, Zhang et al. uncover design principles for engineering the FLS2 immune receptor with broad recognition spectra to detect evasive bacterial pathogens, offering a promising strategy to improve crop disease resistance.

|
Scooped by
The Sainsbury Lab
July 21, 2025 8:40 AM
|
Variation in shoot architecture, or tillering, is an important adaptive trait targeted during the domestication of crops. A well-known regulatory factor in shoot architecture is TEOSINTE BRANCHED 1 (TB1). TB1 and its orthologs have a conserved function in integrating environmental signals to regulate axillary branching or tillering in cereals. The barley (Hordeum vulgare) ortholog of TB1, VULGARE ROW-TYPE SIX 5 (VRS5), regulates tillering and is involved in regulating row-type by inhibiting lateral spikelet development. These discoveries predominantly come from genetic studies; however, how VRS5 regulates these processes on a molecular level remains largely unknown. By combining transcriptome analysis between the vrs5 mutant and the wild type at different developmental stages and DAP-sequencing to locate the genome-wide DNA binding sites of VRS5, we identified bona fide targets of VRS5. We found that VRS5 targets abscisic acid-related genes, potentially to inhibit tillering in a conserved way. Later in inflorescence development, VRS5 also targets row-type gene VRS1 and several known floral development genes, such as MIKCc-type MADS-box genes. This study identifies several genes for mutational analysis, representing a selection of bona fide targets that will contribute to a deeper understanding of the VRS5 network and its role in shaping barley development.

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:20 AM
|
Leucine-rich repeat (LRR) receptor kinases (RKs) and receptor proteins (RPs) are important classes of plant pattern recognition receptors (PRRs) activating pattern-triggered immunity. While both classical and AI-based structural approaches have recently provided crucial insights into ligand-LRR-RK binding mechanisms, our understanding of ligand perception by LRR-RPs remains limited. In contrast to LRR-RKs, many LRR-RPs typically embed one or more loopout regions in their extracellular domains that are crucial for functionality. Here, we employed an AI-based approach to reveal a novel ligand-binding mechanism shared by the Arabidopsis LRR-RPs RLP23 and RLP42 – the PRRs for the short peptide ligands nlp20 and pg13, derived from NECROSIS- AND ETHYLENE-INDUCING PEPTIDE 1-like proteins (NLPs) and fungal endopolygalacturonases (PGs), respectively. This mechanism relies on a β-strand interaction with the N-terminal part of the island domain (ID) loopout, which adopts an antiparallel β-sheet conformation. Additionally, we investigated the larger and more complex binding interface of RLP32 – the PRR for proteobacterial TRANSLATION INITIATION FACTOR 1 (IF1), a folded protein ligand that requires its tertiary structure for recognition. Finally, we describe a mechanistic role of the ID for co-receptor recruitment conserved across LRR-RPs. Together, our results shed light on the ligand-binding mechanisms and receptor complex formation of this important class of PRRs. This opens avenues for a molecular understanding of the plant-pathogen co-evolution, as well as the engineering of plant immune receptors for crop disease resistance.

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:15 AM
|
Effector-triggered immunity (ETI) is a central component of host defense, but whether all cell types execute ETI similarly remains unknown. We combined chemically imposed immune activation with single-cell transcriptomics to profile ETI responses across all leaf cell types in Arabidopsis. Despite uniform ETI perception, we find striking divergence between transcriptional outputs: a core set of defense genes is broadly induced, while distinct cell types activate specialized immune modules. We infer that downstream immune execution is shaped not only by immune receptor activation, but also by cell identity and its associated transcriptional regulatory context, including local transcription factor availability and chromatin accessibility. We further demonstrate that transcriptional regulators preferentially induced in epidermal cells are required to restrict invasion by non-adapted pathogens. Their absence permits pathogen entry into deeper tissues despite intact recognition, revealing a spatial division of immune functions. Our findings uncover a layered immune architecture in plants, challenges the assumption of uniform immune execution, and provides a framework for exploring cell-type-specific resistance logic in multicellular hosts.

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:06 AM
|
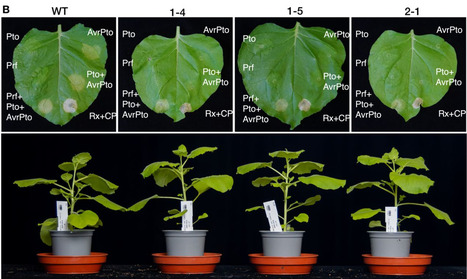
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) can function in networks of sensors and helpers to induce hypersensitive cell death and immunity against pathogens. The tomato sensor NLR Prf guards the Pto kinase from AvrPto and AvrPtoB effector perturbation and activates the downstream helpers NRC2 and NRC3. Prf is conserved across the Solanaceae and its ortholog in the model species Nicotiana benthamiana is also required for detection of AvrPto/AvrPtoB function on Pto. A recent study reported that cell death induction after transient expression of an autoactive mutant of tomato NRC3 is abolished upon RNAi silencing of Prf in N. benthamiana. Here we generated loss-of-function prf mutants in N. benthamiana and demonstrate that autoactive mutants of eight canonical tomato NRCs (NRC0, NRC1, NRC2, NRC3, NRC4a, NRC4b, NRC6, and NRC7) still induce hypersensitive cell death when expressed transiently in the prf mutant background. Autoactive tomato NRCs also triggered cell death when expressed in lettuce (Lactuca sativa), an Asteraceae plant that does not have a Prf ortholog. These results confirm a unidirectional dependency of sensors and helpers in the NRC network and underscore the value of the N. benthamiana and lettuce model systems for studying functional relationships between paired and networked NLRs.

|
Scooped by
The Sainsbury Lab
July 16, 2025 10:59 AM
|
Advances in single-cell and spatial omics technologies have revolutionised biology by revealing the diverse molecular states of individual cells and their spatial organization within tissues. The field of plant biology has widely adopted single-cell transcriptome and chromatin accessibility profiling and spatial transcriptomics, which extend traditional cell biology and genomics analyses and provide unique opportunities to reveal molecular and cellular dynamics of tissues. Using these technologies, comprehensive cell atlases have been generated in several model plant species, providing valuable platforms for discovery and tool development. Other emerging technologies related to single-cell and spatial omics, such as multiomics, lineage tracing, molecular recording, and high-content genetic and chemical perturbation phenotyping, offer immense potential for deepening our understanding of plant biology yet remain underutilised due to unique technical challenges and resource availability. Overcoming plant-specific barriers, such as cell wall complexity and limited antibody resources, alongside community-driven efforts in developing more complete reference atlases and computational tools, will accelerate progress. The synergy between technological innovation and targeted biological questions is poised to drive significant discoveries, advancing plant science. This review highlights the current applications of single-cell and spatial omics technologies in plant research and introduces emerging approaches with the potential to transform the field.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:56 AM
|
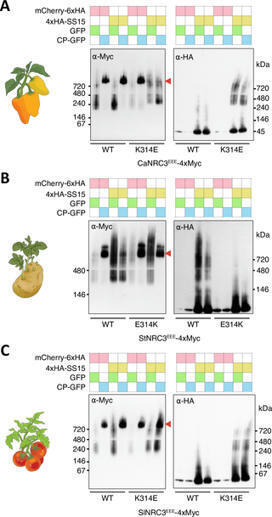
Author summary Plants have immune systems that protect them from various pathogens, including bacteria, fungi and nematodes. This immune system harnesses proteins called NLRs (nucleotide-binding and leucine-rich repeat proteins) to detect and respond to pathogens. However, pathogens often evolve strategies to suppress plant immunity. For example, the cyst nematode Globodera rostochiensis produces an effector protein called SPRYSEC15 (SS15) that inhibits certain NLR proteins, suppressing the plant’s immune response. In this study, we explored how a group of NLR proteins called NRCs, found in the Solanaceae plant family, have evolved to counteract SS15 inhibition. We discovered that while SS15 inhibits all tested NRC2 orthologs, some natural variants of NRC1 and NRC3 are insensitive to its effects. By reconstructing the evolutionary history of NRC3, we found that these proteins acquired insensitivity to SS15 through a specific amino acid substitution about 19 million years ago. Our findings highlight a previously understudied type of evolutionary arms race between plants and pathogens, where plants adapt to evade pathogen-derived suppressors. This research expands our understanding of plant-pathogen coevolution and offers insights that could guide the development of crops with improved disease resistance.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:54 AM
|
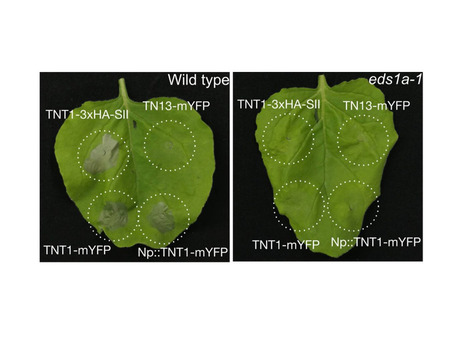
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors detect pathogen-secreted effectors inside host cells and induce a robust immune response, typically involving hypersensitive cell death. NLR genes are often genetically linked and can function as pairs or within larger NLR networks. We previously showed that the truncated Toll/Interleukin-1 Receptor (TIR)-type NLR TIR-NB13 (TN13) is required for resistance of Arabidopsis to Pseudomonas syringae pv. tomato (Pst) DC3000 lacking the type-III effector proteins AvrPto and AvrPtoB. TN13 is genetically linked to a full length TIR-NB-LRR (TNL) gene on chromosome 3. Here, we show that TN13, and its genetically linked TNL both localize to the ER membrane via N-terminal transmembrane domains, are required for resistance to Pst DC3000 (ΔAvrPto/AvrPtoB) and interact with each other in transient expression assays in Nicotiana benthamiana. In contrast to TN13, the full length TNL, which we named TN13-INTERACTING TNL1 (TNT1), induces an autoactive cell death response when expressed in N. benthamiana that depends on an atypical MHV motif in its NB-ARC domain, as well as the EDS1/SAG101/NRG1 module. TN13 and TNT1 furthermore interact with phylogenetically related NLRs encoded by a segmentally duplicated region on chromosome 5. Our data suggest that both TN13 and the genetically linked TNT1 could be part of a larger TIR-type NLR immune regulatory network, in which TNT1 contributes to basal immunity and might function as an autoactive death switch to induce cell death upon pathogen detection.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:51 AM
|
Phakopsora pachyrhizi, an obligate biotrophic rust fungus, is the causal agent of Asian Soybean Rust (ASR) disease. Here, we utilized whole-genome dat…

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:46 AM
|
The receptor-like cytoplasmic kinase BIK1 and its close homolog PBL1 have been widely recognized as central components of plant immunity. However, most genetic studies of BIK1 and PBL1 functions were carried out with single T-DNA insertional mutant alleles. Some phenotypes observed in these mutants, e.g. autoimmunity, have been difficult to reconcile with the proposed role of BIK1 and PBL1 in pattern-triggered immunity. In this study, we generated multiple new alleles of bik1 and pbl1 by CRISPR-Cas9-based gene editing and systematically analyzed these mutants alongside existing T-DNA insertional lines. These analyses reinforced the central role of BIK1 and PBL1 in pattern-triggered immunity mediated by both receptor kinases and receptor-like proteins. At the same time, however, we revealed several pleiotropic phenotypes associated with T-DNA insertions that are not necessarily linked to loss of BIK1 or PBL1 function. Further analyses of newly generated bik1 pbl1 double mutants uncovered an even greater contribution of these kinases to immune signaling and disease resistance than previously appreciated. These findings clarify longstanding ambiguities surrounding BIK1 and PBL1 functions.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:41 AM
|
Plant nucleotide-binding, leucine-rich-repeat (NLR) immune receptors recognize pathogen effectors and activate immunity. The NLR RPS2 recognizes AvrRpt2, a Pseudomonas effector that promotes virulence by proteolytically cleaving a membrane-tethered host protein, RIN4. RIN4 cleavage by AvrRpt2 generates fragments that activate RPS2. A model for RPS2 activation by RIN4 destruction is consistent with the ectopic activity of RPS2 in plants lacking RIN4 but does not explain the link between AvrRpt2’s virulence activity and RPS2 activation. We found that non-membrane-tethered RIN4 derivatives are potent cytosolic activators of RPS2. Activation of RPS2 by these RIN4 derivatives, like AvrRpt2-induced activation, and unlike ectopic activation in the absence of RIN4, requires the defense signaling protein NDR1. Cleavage products of RIN4 produced by AvrRpt2 play contrasting roles in the activation of RPS2, with the membrane-tethered C-terminal fragment suppressing RPS2 and the non-membrane-tethered internal fragment, dependent on compatibility with the C-terminal fragment, overcoming its suppression of RPS2.
|

|
Scooped by
The Sainsbury Lab
August 18, 2025 6:10 AM
|
Protein evolution is influenced by historical contingencies and functional constraints, but their combined impact on rapidly diversifying pathogen virulence effectors remains poorly understood. Here, we combined ancestral state reconstructions and functional assays to recapitulate the evolution of the MAX-fold effector protein APikL2 of the plant pathogenic blast fungus Magnaporthe (syn. Pyricularia) oryzae, focusing on the ancestral and functionally critical amino acid residue D66 (Asp, Codon: GAT). “Rewinding the tape” experiments based on ancestral sequence resurrection revealed that, out of the seven potential amino acid substitutions derived from single nucleotide polymorphisms, only the naturally occurring D66N (Asp to Asn, GAT to AAT) expanded the binding spectrum to host plant proteins of the heavy metal associated (HMA) family. In contrast, three of the nonsynonymous substitutions were deleterious resulting in loss of binding to HMA proteins. Additionally, we identified three cases of homoplasy in the APikL effector family, involving HMA-binding interfaces, indicating recurrent convergent evolution. Our findings suggest an experimental framework for predicting evolutionary outcomes of pathogen effector—host target interactions with implications for plant disease resistance breeding.

|
Scooped by
The Sainsbury Lab
July 31, 2025 5:37 AM
|

|
Scooped by
The Sainsbury Lab
July 22, 2025 12:24 PM
|
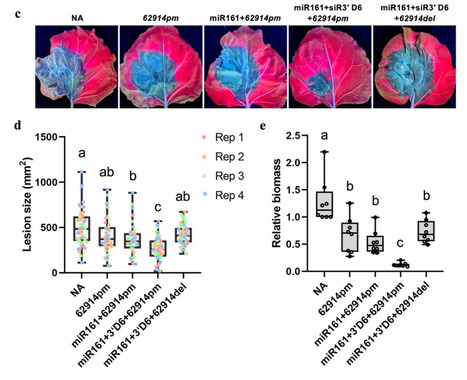
Small RNA-mediated gene silencing contributes to plant immunity. The secondary small interfering RNA (siRNA) pathway promotes defense by silencing target genes in invading fungal and oomycete pathogens. Many secondary siRNAs derive from transcripts potentially encoding pentatricopeptide repeat (PPR) proteins. Here, we report that siRNA production is an ancient function of an evolutionarily conserved clade of PPR genes that undergo extensive within-species diversification. In Arabidopsis thaliana, siRNA-source PPRs are physically clustered in one locus on Chromosome 1. These sequences are diversified through gene duplication followed by sequence diversification as well as accumulation of high-impact variations including pseudogenization. This diversity leads to the accumulation of a diverse PPR-siRNA pool, consistent with an engagement in a co-evolutionary arms race with the pathogens. This study defines siRNA-producing PPRs as a family of defense genes and highlights the potential of PPR-siRNA-based engineering for enhancing broad-spectrum disease resistance.

|
Scooped by
The Sainsbury Lab
July 18, 2025 3:55 AM
|
Nucleotide binding and leucine-rich repeat (NLR) proteins are intracellular immune receptors that occur across all kingdoms of life but are particularly highly diversified in plants (Barragan & Weigel, 2021). In plants, NLRs that carry a coiled-coil (CC) domain at their N termini are the most phylogenetically widespread class. Following pathogen recognition, CC-NLR proteins oligomerize into pentameric or hexameric pore-like complexes (Wang et al., 2019; Förderer et al., 2022; Zhao et al., 2022; Liu et al., 2024; Madhuprakash et al., 2024). These complexes, known as resistosomes, are a defining feature of NLRs that execute the immune response; some of them are known to translocate to cellular membranes and trigger immune responses such as calcium influx and hypersensitive cell death (Duggan et al., 2021; Contreras et al., 2022; Ibrahim et al., 2024). The prevailing model is that the funnel-shaped structure of CC-NLR resistosomes inserts into membranes and is required for executing the cell death and immune response (Wang et al., 2019; Adachi et al., 2019b; Förderer & Kourelis, 2023). This funnel-shaped structure is formed by the N-terminal α1 helix, which is a structurally dynamic region that is difficult to resolve using cryo-electron microscopy (cryo-EM) (Förderer et al., 2022; Zhao et al., 2022; Liu et al., 2024; Madhuprakash et al., 2024).

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:19 AM
|
Pathogens counteract central nodes of NLR immune receptor networks to suppress immunity. However, the mechanisms by which pathogens hijack helper NLR pathways are poorly understood. Here, we show that an effector from the potato late blight pathogen Phytophthora infestans bridges the host protein NbTOL9a, a putative member of the host ESCRT pathway, to a helper NLR to suppress immunity. In this work, we solved the crystal structure of the RXLR-LWY effector AVRcap1b in complex with the ENTH domain of NbTOL9a. The structure revealed that unlike other RXLR-LWY effectors, AVRcap1b has a novel L-shaped fold that defines a new structural family of effectors in the Phytophthora genus. Moreover, we defined the AVRcap1b/NbTOL9a binding interface and designed effector mutants that don’t bind NbTOL9a, impairing immune suppression. This indicates that ENTH binding is required for full virulence activity of this effector. Lastly, we show that AVRcap1b associates specifically with activated NbNRC2 independently of NbTOL9a binding. This suggests that the effector functions as a bridge that interconnects NbNRC2 with the NbTOL9a pathway. These results illustrate an unprecedented pathogen mechanism to hijack helper NLR pathways and suppress immunity.

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:13 AM
|
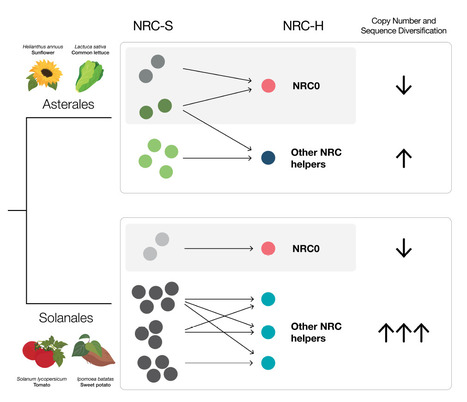
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) are known for their rapid evolution, even at the intraspecific level, yet the rates of evolution differ significantly across various NLR classes. Within the NRC (NLR Required for Cell Death) network, NLRs operate in complex sensor-helper configurations to confer immunity against a diverse array of pathogens, particularly in Asterids. While helper NLRs are typically conserved and evolve slowly, sensor NLRs tend to evolve more rapidly. However, the functional connections between slow and fast-evolving NLRs remain poorly understood, notably in important crop species. We conducted a comparative analysis of NLRs across 40 Solanales and 29 Asterales genomes to explore NRC network expansion and diversification within the less-studied Asterales order. Our findings reveal that the NRC network has expanded less in Asterales compared to Solanales. We functionally validated a minimal Asterales NRC network with 2 helpers and 9 sensors in common lettuce (Lactuca sativa). Through selection and diversification analysis and structural modeling of NRC helper and sensor subclades in the Lactuca genus, we found varying evolutionary diversification rates between NRC helpers and sensors. We found a correlation between sensor diversification rates and helper dependency, with sensors reliant on a phylogenetically conserved helpers experiencing limited diversification pressure. Our results highlight the lineage- and function-specific evolution of the NRC network, offering insights into the evolutionary pressures shaping plant immune receptor networks.

|
Scooped by
The Sainsbury Lab
July 16, 2025 11:04 AM
|
- Phytoplasmas are specialized phloem-limited bacteria that cause diseases on various crops, resulting in significant agricultural losses. This research focuses on the jujube witches' broom (JWB) phytoplasma and investigates the host-manipulating activity of the effector SJP39.
- We found that SJP39 directly interacts with the plant transcription factor bHLH87 in the nuclei. SJP39 stabilizes the bHLH87 homologs in Arabidopsis thaliana and jujube, leading to growth defects in the plants.
- Transcriptomic analysis indicates that SJP39 affects the gibberellin (GA) pathway in jujube. We further demonstrate that ZjbHLH87 regulates GA signaling as a negative regulator, and SJP39 enhances this regulation.
- The research offers important insights into the pathogenesis of JWB disease and identified SJP39 as a virulence factor that can contribute to the growth defects caused by JWB phytoplasma infection. These findings open new opportunities to manage JWB and other phytoplasma diseases.

|
Scooped by
The Sainsbury Lab
July 16, 2025 10:54 AM
|
The rice blast fungus Magnaporthe oryzae secretes a battery of effector proteins to facilitate host infection. Among these effectors, pathogenicity toward weeping lovegrass 2 (Pwl2) was originally identified as a host specificity determinant for the infection of weeping lovegrass (Eragrostis curvula) and is also recognized by the barley (Hordeum vulgare) Mla3 resistance protein. However, the biological activity of Pwl2 remains unknown. Here, we showed that the Pmk1 MAP kinase regulates PWL2 expression during the cell-to-cell movement of M. oryzae at plasmodesmata-containing pit fields. Consistent with this finding, we provided evidence that Pwl2 binds to the barley heavy metal–binding isoprenylated protein HIPP43, which results in HIPP43 displacement from plasmodesmata. Transgenic barley lines overexpressing PWL2 or HIPP43 exhibit attenuated immune responses and increased disease susceptibility. In contrast, a Pwl2SNDEYWY variant that does not interact with HIPP43 fails to alter the plasmodesmata localization of HIPP43. Targeted deletion of 3 PWL2 copies in M. oryzae resulted in a Δpwl2 mutant showing gain of virulence toward weeping lovegrass and barley Mla3 lines, but reduced blast disease severity on susceptible host plants. Taken together, our results provide evidence that Pwl2 is a virulence factor that suppresses host immunity by perturbing the plasmodesmatal deployment of HIPP43.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:55 AM
|
The Arabidopsis TIR-NLR immune receptors RPS4 and RRS1 function together to enable recognition of multiple effector proteins including AvrRps4 and PopP2. We show here that both in the presence and absence of effector, RPS4 and RRS1 form an oligomer that does not change in size upon effector provision. Oligomer formation involves interactions between the RPS4 and RRS1 TIR domains and requires nucleotide binding capacity in RPS4. RPS4 mutants that lose TIR domain NADase activity abrogate immune activation but retain oligomerization. A cysteine residue in the RPS4 LRR domain contributes to oligomer stabilization. We propose that upon effector recognition, conformational changes in the complex relieve inhibition of RPS4 TIR domains by RRS1 TIR domains, enabling proximity between RPS4 TIR domains to create NADase activity.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:53 AM
|
Dioecious plants, which have distinct male and female individuals, constitute ∼5% of angiosperm species and have emerged frequently and independently from hermaphroditic ancestors. Although recent molecular studies of sex determination have started to reveal the diversity of the genetic systems underlying dioecy, research on the evolution of dioecy is limited, especially in monocots. Here, we describe the molecular basis of sex determination in the monocot Dioscorea tokoro, a dioecious wild yam endemic to East Asia. Chromosome-scale and haplotype-resolved genome assemblies and linkage analysis suggested that this plant has a male heterogametic sex determination (XY) system, with sex determination regions located on chromosome 3. Sequence read coverage analysis of the sex chromosomes revealed X- and Y-specific regions in putative pericentromeric chromosome regions. Within the Y-specific region, we identified two candidate genes that are likely involved in sex determination: BLH9, encoding a homeobox protein, and HSP90, encoding a molecular chaperone. BLH9 has similar functions to AtBLH9 in Arabidopsis thaliana. BLH9 is thought to suppress female organ development, whereas HSP90 might be required for pollen development. These results shed light on the complex evolution of dioecy in plants.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:48 AM
|
Plants constantly monitor their environment to adapt to potential threats to their health and fitness. This involves cell-surface receptors that can detect conserved microbe-associated molecular patterns (MAMPs) or endogenous immunogenic signals, initiating signaling pathways to induce broad-spectrum disease resistance, known as pattern-triggered immunity (PTI). In Arabidopsis thaliana, the leucine-rich repeat receptor kinase (LRR-RK) MIK2 is an exceptionally versatile receptor involved in the perception of the vast family of Brassicales-specific endogenous SCOOP peptides as well as potential MAMPs derived from Fusarium and related fungi. Although only plant species belonging to the order of Brassicales encode genes for SCOOP peptides and show SCOOP-responsiveness, the Fusarium-derived elicitor fraction also induces PTI responses in plants from other lineages. In this study, we demonstrate that Fusarium elicitor-responsiveness and proteins belonging to the MIK2-clade are widely conserved among seed plants. We identified a MIK2-clade protein from tomato, which shares properties of AtMIK2 in the perception of the Fusarium elicitor but not of SCOOP peptides. Tomato mutants lacking the receptor show compromised PTI responses to the fungal elicitor and enhanced susceptibility to infection by Fusarium oxysporum. Our data provide insights into the evolutionary trajectory of MIK2 as a multifunctional receptor involved in plant immunity.

|
Scooped by
The Sainsbury Lab
July 16, 2025 5:43 AM
|
Our usual encounters with fungi are when we observe mushrooms in forests or moulds on food that we failed to eat on time. In both instances, however, we are only seeing a very limited view of fungal growth. Fungi are osmotrophs, which means that they consume the food that surrounds them by secreting enzymes to degrade polymers into simple sugars, fatty acids and amino acids. This type of growth has a number of consequences - it explains why fungi secrete toxins and antibiotics to protect their food sources from competitors and why they can grow so rapidly. It also explains why fungi have evolved the capacity to forcefully invade hard substrates, such as wood, animal skins and the cuticles of plants. By doing so, they can access new sources of food, often inaccessible to their competition, and this has enabled fungi to become highly successful pathogens of both animals and plants, causing diseases in organisms as diverse as insects, amphibians, humans, reptiles, and rice plants. It is becoming clear that, in addition to their prodigious secretion of enzymes to degrade complex substrates, fungi can exert very substantial physical forces. Such forces are probably essential for many aspects of the fungal lifestyle, including colonisation of their usual habitats like soil and leaf litter, which require penetration and invasion to enable their digestion by fungi. But for pathogenic fungi, the requirement for invasive growth is even more acute. In this Primer, we explore the mechanobiology of fungal invasive growth and the emerging view of the different mechanisms that fungal pathogens deploy to gain entry to host tissue. We focus mainly on plant pathogens, where recent experimental work has been most extensive, and highlight key research questions for the future.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...