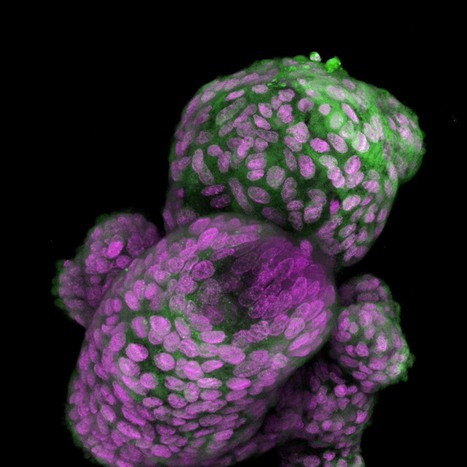
Neuromodulation technologies are crucial for investigating neuronal connectivity and brain function. Magnetic neuromodulation offers wireless and remote deep brain stimulations that are lacking in optogenetic- and wired-electrode-based tools. However, due to the limited understanding of working principles and poorly designed magnetic operating systems, earlier magnetic approaches have yet to be utilized. Furthermore, despite its importance in neuroscience research, cell-type-specific magnetic neuromodulation has remained elusive. Here we present a nanomaterials-based magnetogenetic toolbox, in conjunction with Cre-loxP technology, to selectively activate genetically encoded Piezo1 ion channels in targeted neuronal populations via torque generated by the nanomagnetic actuators in vitro and in vivo. We demonstrate this cell-type-targeting magnetic approach for remote and spatiotemporal precise control of deep brain neural activity in multiple behavioural models, such as bidirectional feeding control, long-term neuromodulation for weight control in obese mice and wireless modulation of social behaviours in multiple mice in the same physical space. Our study demonstrates the potential of cell-type-specific magnetogenetics as an effective and reliable research tool for life sciences, especially in wireless, long-term and freely behaving animals. Minimally invasive cellular-level target-specific neuromodulation is needed to decipher brain function and neural circuitry. Here nano-magnetogenetics using magnetic force actuating nanoparticles has been reported, enabling wireless and remote stimulation of targeted deep brain neurons in freely behaving animals.






 Your new post is loading...
Your new post is loading...













In a recent study published on the bioRxiv preprint server, researchers evaluated the impact of gene amplification on cross-species adaptation. The researchers demonstrated that the impact of RhTRS1 gene amplification could effectively rescue viral replication in fibroblasts from partially resistant African green monkeys (AGMs). The team infected A549 cells with vaccinia virus (VACV) + RhTRS1 or the VACV strain named Copenhagen, expressing a beta-galactosidase (bg) reporter gene. The team explored whether amplification of rhtrs1 could increase the extent of viral replication in the absence of PKR. The team also infected RNase L competent A549 cells or regularly spaced short palindromic repeats with CRISPR in clusters of RNase L deletion present in existing A549 cells with VACV-RhTRS1, VACV-bg or AGM-A. The results of the study showed that A549 cells restricted VACV + RhTRS1 replication by nearly 10,000-fold compared to VACV-bg replication. The team noted that deletion of A549 PKR cells enhanced VACV + RhTRS1 replication by up to 1000-fold compared to PKR-competent cells. The team observed that RNAase L had no impact on VACV-bg replication.