 Your new post is loading...

|
Scooped by
?
March 9, 2018 3:36 PM
|
The molybdenum nitrogenase is responsible for most biological nitrogen fixation, a prokaryotic metabolic process that determines the global biogeochemical cycles of nitrogen and carbon. Here we describe the trafficking of molybdenum for nitrogen fixation in the model diazotrophic bacterium Azotobacter vinelandii. The genes and proteins involved in molybdenum uptake, homeostasis, storage, regulation, and nitrogenase cofactor biosynthesis are reviewed. Molybdenum biochemistry in A. vinelandii reveals unexpected mechanisms and a new role for iron-sulfur clusters in the sequestration and delivery of molybdenum.

|
Scooped by
?
March 8, 2018 5:40 PM
|
Symbiotic nitrogen fixation in legume root nodules requires a steady supply of molybdenum for synthesis of the iron-molybdenum cofactor of nitrogenase. This nutrient has to be provided by the host plant from the soil, crossing several symplastically disconnected compartments through molybdate transporters, including members of the MOT1 family. MtMOT1.2 is a Medicago truncatula MOT1 family member located in the endodermal cells in roots and nodules. Immunolocalization of a tagged MtMOT1.2 indicates that it is associated to the plasma membrane and to intracellular membrane systems, where it would be transporting molybdate towards the cytosol, as indicated in yeast transport assays. A loss-of-function mot1.2-1 mutant showed reduced growth compared to wild-type plants when nitrogen fixation was required, but not when nitrogen was provided as nitrate. While no effect on molybdenum-dependent nitrate reductase activity was observed, nitrogenase activity was severely affected, explaining the observed difference of growth depending on nitrogen source. This phenotype was the result of molybdate not reaching the nitrogen-fixing nodules, since genetic complementation with a wild-type MtMOT1.2 gene or molybdate-fortification of the nutrient solution, both restored wild-type levels of growth and nitrogenase activity. These results support a model in which MtMOT1.2 would mediate molybdate delivery by the vasculature into the nodules.

|
Scooped by
?
March 8, 2018 12:39 PM
|
RNA occupies a sweeping middle ground in molecular and cellular biology. Its cousin, DNA, is far easier to study and sequence; it is more chemically stable and folds into more consistent secondary and tertiary structures. But much of the information about a cell’s activity is embedded in its RNA sequences—in the isoforms transcribed, in splicing patterns, and in the function of noncoding sequences. Researchers are just scratching the surface of the biology in the transcriptome and its implications for both normal cellular function and disease pathology. One critical challenge with today’s RNA sequencing is trying to balance experimental goals with technology limitations. Illumina’s systems remain the gold standard for nucleic acid sequencing. According to Vollmers, this sequencing strategy is fantastic for many applications, including quantifying gene expression and finding individual splice junctions, and it has unparalleled accuracy because of the volume of reads. But read length is a limiting factor. Short-read sequencing can be a bit like trying to reconstruct paragraphs of text word by word—a daunting process. By contrast, long-read sequencing is like recording an entire paragraph instead, said Hagen Tilgner of Weill Cornell Medical College. With that context, it’s far easier to make overall sense of a transcript.

|
Scooped by
?
March 7, 2018 12:53 PM
|
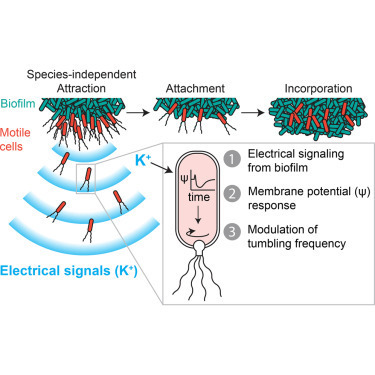
• Electrical signaling within biofilms attracts distant motile cells • Attraction is caused by membrane-potential-dependent modulation of tumbling frequency • Electrical signaling is generic, resulting in species-independent attraction • Attraction leads to incorporation of diverse species into a pre-existing biofilm
Bacteria residing within biofilm communities can coordinate their behavior through cell-to-cell signaling. However, it remains unclear if these signals can also influence the behavior of distant cells that are not part of the community. Using a microfluidic approach, we find that potassium ion channel-mediated electrical signaling generated by a Bacillus subtilis biofilm can attract distant cells. Integration of experiments and mathematical modeling indicates that extracellular potassium emitted from the biofilm alters the membrane potential of distant cells, thereby directing their motility. This electrically mediated attraction appears to be a generic mechanism that enables cross-species interactions, as Pseudomonas aeruginosa cells also become attracted to the electrical signal released by the B. subtilis biofilm. Cells within a biofilm community can thus not only coordinate their own behavior but also influence the behavior of diverse bacteria at a distance through long-range electrical signaling.

|
Scooped by
?
March 7, 2018 9:27 AM
|
Metabolism is one of the best‐understood cellular processes whose network topology of enzymatic reactions is determined by an organism's genome. The influence of genes on metabolite levels, however, remains largely unknown, particularly for the many genes encoding non‐enzymatic proteins. Serendipitously, genomewide association studies explore the relationship between genetic variants and metabolite levels, but a comprehensive interaction network has remained elusive even for the simplest single‐celled organisms. Here, we systematically mapped the association between > 3,800 single‐gene deletions in the bacterium Escherichia coli and relative concentrations of > 7,000 intracellular metabolite ions. Beyond expected metabolic changes in the proximity to abolished enzyme activities, the association map reveals a largely unknown landscape of gene–metabolite interactions that are not represented in metabolic models. Therefore, the map provides a unique resource for assessing the genetic basis of metabolic changes and conversely hypothesizing metabolic consequences of genetic alterations. We illustrate this by predicting metabolism‐related functions of 72 so far not annotated genes and by identifying key genes mediating the cellular response to environmental perturbations.

|
Scooped by
?
March 7, 2018 1:25 AM
|
The aim of this dataset is to identify and collect compounds that are known for being detectable by a living cell, through the action of a genetically encoded biosensor and is centred on bacterial transcription factors. Such a dataset should open the possibility to consider a wide range of applications in synthetic biology. The reader will find in this dataset the name of the compounds, their InChI (molecular structure), the publication where the detection was reported, the organism in which this was detected or engineered, the type of detection and experiment that was performed as well as the name of the biosensor. A comment field is also provided that explains why the compound was included in the dataset, based on quotes from the reference publication or the database it was extracted from. Manual curation of ACS Synthetic Biology abstracts (Volumes 1 to 6 and Volume 7 issue 1) was performed as well as extraction from the following databases: Bionemo v6.0 (Carbajosa et al., 2009) [1], RegTransbase r20120406 (Cipriano et al., 2013) [2], RegulonDB v9.0 (Gama-Castro et al., 2016) [3], RegPrecise v4.0 (Novichkov et al., 2013) [4] and Sigmol v20180122 (Rajput et al., 2016) [5].

|
Scooped by
?
March 6, 2018 1:25 PM
|
In efforts to translate basic-science results into pharmaceuticals and other technologies, success cannot be taken for granted.

|
Scooped by
?
March 2, 2018 11:21 AM
|
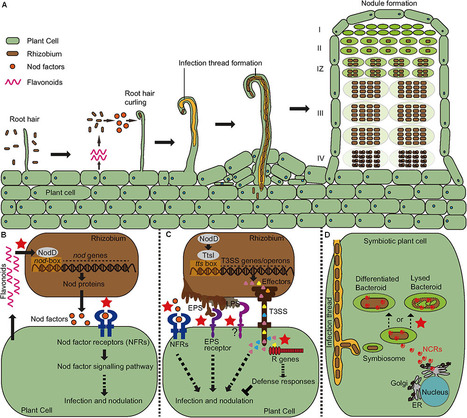
Legumes are able to form a symbiotic relationship with nitrogen-fixing soil bacteria called rhizobia. The result of this symbiosis is to form nodules on the plant root, within which the bacteria can convert atmospheric nitrogen into ammonia that can be used by the plant. Establishment of a successful symbiosis requires the two symbiotic partners to be compatible with each other throughout the process of symbiotic development. However, incompatibility frequently occurs, such that a bacterial strain is unable to nodulate a particular host plant or forms nodules that are incapable of fixing nitrogen. Genetic and molecular mechanisms that regulate symbiotic specificity are diverse, involving a wide range of host and bacterial genes and signals with various modes of action. In this review, we will provide an update on our current knowledge of how the recognition specificity has evolved in the context of symbiosis signaling and plant immunity.

|
Scooped by
?
March 2, 2018 11:05 AM
|
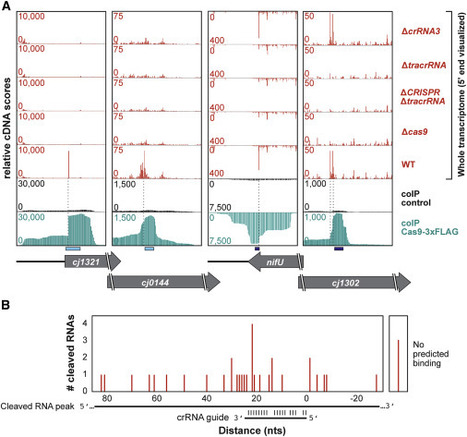
Cas9 nucleases naturally utilize CRISPR RNAs (crRNAs) to silence foreign double-stranded DNA. While recent work has shown that some Cas9 nucleases can also target RNA, RNA recognition has required nuclease modifications or accessory factors. Here, we show that the Campylobacter jejuni Cas9 (CjCas9) can bind and cleave complementary endogenous mRNAs in a crRNA-dependent manner. Approximately 100 transcripts co-immunoprecipitated with CjCas9 and generally can be subdivided through their base-pairing potential to the four crRNAs. A subset of these RNAs was cleaved around or within the predicted binding site. Mutational analyses revealed that RNA binding was crRNA and tracrRNA dependent and that target RNA cleavage required the CjCas9 HNH domain. We further observed that RNA cleavage was PAM independent, improved with greater complementarity between the crRNA and the RNA target, and was programmable in vitro. These findings suggest that C. jejuni Cas9 is a promiscuous nuclease that can coordinately target both DNA and RNA.

|
Scooped by
?
March 2, 2018 9:16 AM
|
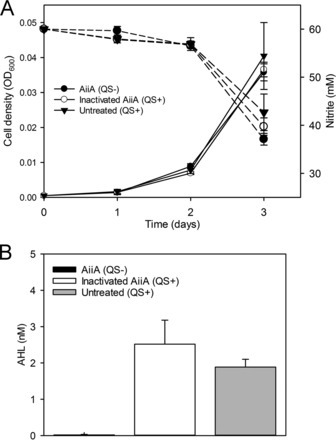
To date, most studies on QS have focused on model bacteria that are amenable to genetic manipulation and capable of high growth rates, but many environmentally important bacteria have been overlooked. For example, representatives of proteobacteria that participate in nitrification, the aerobic oxidation of ammonia to nitrate via nitrite, produce QS signals called acyl-homoserine lactones (AHLs). Nitrification emits nitrogen oxide gases (NO, NO2, and N2O), which are potentially hazardous compounds that contribute to global warming. Despite considerable interest in nitrification, the purpose of QS in the physiology/ecology of nitrifying bacteria is poorly understood. Through a quorum quenching approach, we investigated the role of QS in a well-studied AHL-producing nitrite oxidizer, Nitrobacter winogradskyi. We added a recombinant AiiA lactonase to N. winogradskyi cultures to degrade AHLs to prevent their accumulation and to induce a QS-negative phenotype and then used mRNA sequencing (mRNA-Seq) to identify putative QS-controlled genes. Our transcriptome analysis showed that expression of nirK and nirK cluster genes (ncgABC) increased up to 19.9-fold under QS-proficient conditions (minus active lactonase). These data led to us to query if QS influenced nitrogen oxide gas fluxes in N. winogradskyi. Production and consumption of NOx increased and production of N2O decreased under QS-proficient conditions. Quorum quenching transcriptome approaches have broad potential to identify QS-controlle

|
Scooped by
?
March 1, 2018 11:54 PM
|
The acquisition of a virulence plasmid is sufficient to turn a beneficial strain of Rhodococcus bacteria into a pathogen. "Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management"

|
Scooped by
?
March 1, 2018 9:09 AM
|
Harnessing and controlling self-assembly is an important step in developing proteins as novel biomaterials. With this goal, here we report the design of a general genetically programmed system that covalently concatenates multiple distinct protein domains into specific assembled arrays. It is driven by iterative intein-mediated Native Chemical Ligation (NCL) under mild native conditions. The system uses a series of initially inert recombinant protein fusions that sandwich the protein modules to be ligated between one of a number of different affinity tags and an intein protein domain. Orthogonal activation at opposite termini of compatible protein fusions, via protease and intein cleavage, coupled with sequential mixing directs an irreversible and traceless stepwise assembly process. This gives total control over the composition and arrangement of component proteins within the final product, enabled the limits of the system - reaction efficiency and yield - to be investigated and led to the production of “functional” assemblies.

|
Scooped by
?
March 1, 2018 8:30 AM
|
The compartmentalization of enzymes into organelles is a promising strategy for limiting metabolic crosstalk and improving pathway efficiency; however, prokaryotes are unicellular organisms that lack membrane-bound organelles. To mimic this natural compartmentalization, we present here the targeting of the reductive tricarboxylic acid (rTCA) pathway to the periplasm to enhance the production of malate. A multigene combination knockout strategy was used to construct a phosphoenolpyruvate (PEP) pool. Then, the genes encoding phosphoenolpyruvate carboxykinase and malate dehydrogenase were combinatorially overexpressed to construct a cytoplasmic rTCA pathway for malate biosynthesis; however, the efficiency of malate production was low. To further enhance malate production, the rTCA pathway was targeted to the periplasm, which led to a 100% increase in malate production to 18.8 mM. Next, dual metabolic engineering regulation was adopted to balance the cytoplasmic and periplasmic pathways, leading to an increase in malate production to 58.8 mM. The final engineered strain, GL2306, produced 193 mM malate with a yield of 0.53 mol/mol in 5 L of pH-stat fed-batch culture. The strategy described here paves the way for the development of metabolic engineering and synthetic biology in the microbial production of chemicals. This article is protected by copyright. All rights reserved.
|

|
Scooped by
?
March 8, 2018 11:04 PM
|
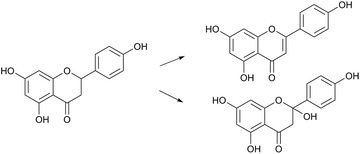
2-Oxoglutarate-dependent oxygenases (2ODOs) comprise a large enzyme superfamily in plant genomes, second in size only to the cytochromes P450 monooxygenase (CYP) superfamily. 2ODOs participate in both primary and specialized plant pathways, and their occurrence across all life kingdoms points to an ancient origin. Phylogenetic evidence supports substantial expansion and diversification of 2ODOs following the split from the common ancestor of land plants. More conserved roles for these enzymes include oxidation within hormone metabolism, such as the recently described capacity of Dioxygenase for Auxin Oxidation (DAO) for governing auxin homeostasis. Conserved structural features among 2ODOs has provided a basis for continued investigation into their mechanisms, and recent structural work is expected to illuminate intriguing reactions such as that of 1-aminocyclopropane-1-carboxylic acid oxidase (ACCO). Phylogenetic radiation among this superfamily combined with neo- and subfunctionalization has enabled recruitment to highly specialized pathways, including those yielding medicines, flavours, dyes, poisons, and compounds important for plant-environment interactions. Catalytic versatility of 2ODOs in plants and across broader taxa continues to inspire biochemists tasked with the discovery of new enzymes. This highlight article summarizes recent reports up to 2018 of 2ODOs within plant metabolism. Furthermore, the respective contributions of 2ODOs and other oxidases to natural product biosynthesis are discussed as a framework for continued discovery.

|
Scooped by
?
March 8, 2018 4:56 PM
|
By virtue of its unique electrochemical properties, iron makes an ideal redox active cofactor for many biologic processes. In addition to its important role in respiration, central metabolism, nitrogen fixation, and photosynthesis, iron also is used as a sensor of cellular redox status. Iron-based sensors incorporate Fe-S clusters, heme, and mononuclear iron sites to act as switches to control protein activity in response to changes in cellular redox balance. Here we provide an overview of iron-based redox sensor proteins, in both prokaryotes and eukaryotes, that have been characterized at the biochemical level. Although this review emphasizes redox sensors containing Fe-S clusters, proteins that use heme or novel iron sites also are discussed.

|
Scooped by
?
March 7, 2018 11:39 PM
|
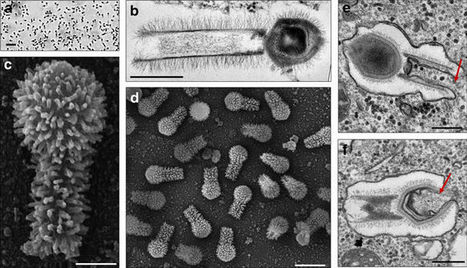
Here we report the discovery of two Tupanvirus strains, the longest tailed Mimiviridae members isolated in amoebae. Their genomes are 1.44–1.51 Mb linear double-strand DNA coding for 1276–1425 predicted proteins. Tupanviruses share the same ancestors with mimivirus lineages and these giant viruses present the largest translational apparatus within the known virosphere, with up to 70 tRNA, 20 aaRS, 11 factors for all translation steps, and factors related to tRNA/mRNA maturation and ribosome protein modification. Moreover, two sequences with significant similarity to intronic regions of 18 S rRNA genes are encoded by the tupanviruses and highly expressed. In this translation-associated gene set, only the ribosome is lacking. At high multiplicity of infections, tupanvirus is also cytotoxic and causes a severe shutdown of ribosomal RNA and a progressive degradation of the nucleus in host and non-host cells. The analysis of tupanviruses constitutes a new step toward understanding the evolution of giant viruses.

|
Scooped by
?
March 7, 2018 12:50 PM
|
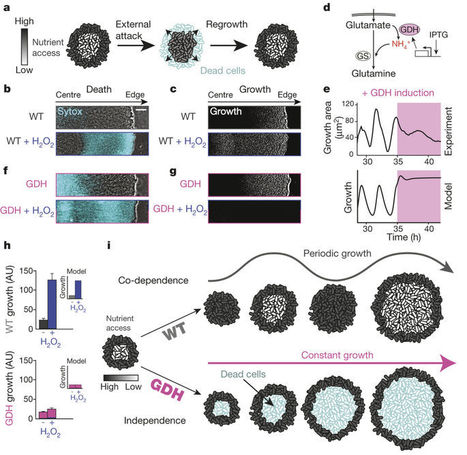
Cells that reside within a community can cooperate and also compete with each other for resources. It remains unclear how these opposing interactions are resolved at the population level. Here we investigate such an internal conflict within a microbial (Bacillus subtilis) biofilm community: cells in the biofilm periphery not only protect interior cells from external attack but also starve them through nutrient consumption. We discover that this conflict between protection and starvation is resolved through emergence of long-range metabolic co-dependence between peripheral and interior cells. As a result, biofilm growth halts periodically, increasing nutrient availability for the sheltered interior cells. We show that this collective oscillation in biofilm growth benefits the community in the event of a chemical attack. These findings indicate that oscillations support population-level conflict resolution by coordinating competing metabolic demands in space and time, suggesting new strategies to control biofilm growth.

|
Scooped by
?
March 7, 2018 9:20 AM
|
We demonstrate the genetic modification of structural color in a living system by using bacteria Iridescent 1 (IR1) as a model system. IR1 colonies consist of rod-shaped bacteria that pack in a dense hexagonal arrangement through gliding and growth, thus interfering with light to give a bright, green, and glittering appearance. By generating IR1 mutants and mapping their optical properties, we show that genetic alterations can change colony organization and thus their visual appearance. The findings provide insight into the genes controlling structural color, which is important for evolutionary studies and for understanding biological formation at the nanoscale. At the same time, it is an important step toward directed engineering of photonic systems from living organisms.
Naturally occurring photonic structures are responsible for the bright and vivid coloration in a large variety of living organisms. Despite efforts to understand their biological functions, development, and complex optical response, little is known of the underlying genes involved in the development of these nanostructures in any domain of life. Here, we used Flavobacterium colonies as a model system to demonstrate that genes responsible for gliding motility, cell shape, the stringent response, and tRNA modification contribute to the optical appearance of the colony. By structural and optical analysis, we obtained a detailed correlation of how genetic modifications alter structural color in bacterial colonies. Understanding of genotype and phenotype relations in this system opens the way to genetic engineering of on-demand living optical materials, for use as paints and living sensors.

|
Scooped by
?
March 6, 2018 11:55 PM
|
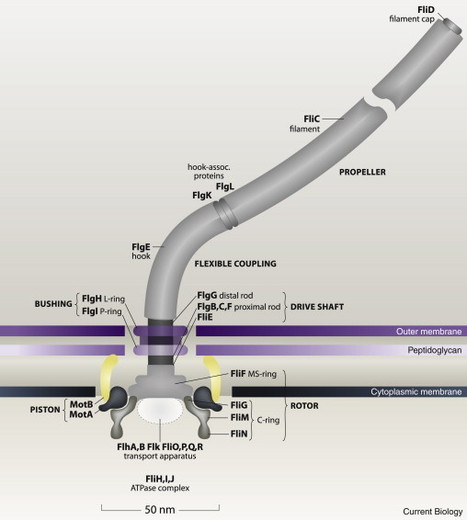
How does the filament generate thrust? The viscous drag on a thin stick in water is about twice as large when it moves sideways than when it moves lengthwise. Propulsion depends upon this asymmetry. In common bacteria, the normal filament is long and thin: the pitch of the helix is about five times its diameter. Think of this helix as a series of sticks moving slantwise in the aqueous medium, rotate the helix about its long axis, and add-up the forces due to the viscous drag on each stick. You will find that the helix generates both thrust and torque (axial force and circumferential twist). Suppose you hold such a helix in front of you and turn it CW: it will push you backwards and try to twist you CCW. An observer looking at you from the far end of the helix will see the filament rotate CCW, your body rotate more slowly CW, and the pair of you move off into the distance. But to do this experiment in a manner relevant to a bacterium, you would have to immerse yourself in a medium much more viscous than water, such as molasses.

|
Scooped by
?
March 2, 2018 1:03 PM
|
A Gram-positive, fast-growing, endophytic bacterium was isolated from root nodules of Medicago polymorpha and identified as Bacillus megaterium. The isolate, named NMp082, co-inhabited nodules with the symbiotic rhizobium Ensifer medicae. B. megaterium NMp082 contained nifH and nodD genes that were 100% identical to those of Ensifer meliloti, an unusual event that suggested previous lateral gene transfer from a different rhizobial species. Despite the presence of nodulation and nitrogen fixation genes, the endophyte was not able to form effective nodules; however, it induced nodule-like unorganised structures in alfalfa roots. Axenic inoculation promoted plant growth in M. polymorpha, Medicago lupulina, Medicago truncatula and Medicago sativa, and co-inoculation with E. medicae enhanced growth and nodulation of Medicago spp. plants compared with inoculation with either bacterium alone. B. megaterium NMp082 also induced tolerance to salt stress in alfalfa and Arabidopsis plants. The ability to produce indole acetic acid (IAA) and the 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity displayed by the endophyte in vitro might explain the observed plant growth promotion and salt stress alleviation. The isolate was also highly tolerant to salt stress, water deficit and to the presence of different heavy metals. The newly characterised endophytic bacterium possessed specific characteristics that point at potential applications to sustain plant growth and nodulation under abiotic stress.

|
Scooped by
?
March 2, 2018 11:17 AM
|
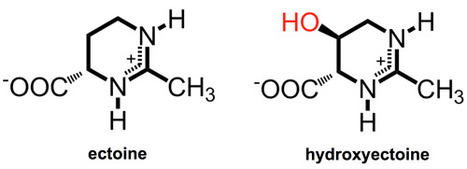
The ability to maintain physiologic and metabolic functions in response to changes in the salinity of the surrounding environment is critical for microorganisms. These organisms have adopted two strategies for dealing with fluctuations in environmental salinity: “salt in,” which occurs when the cells encounter hyperosmotic conditions, and “salt out.” A signature of organisms that use the “salt in” strategy is an acidic proteome that favors persistent high relative concentrations of cations. Organisms that use the “salt out” mechanism also import potassium in response to hyperosmotic shock; however, they do not maintain it for extended periods. Instead, they either import or synthesize compatible solutes that include ectoine and glycine-betaine

|
Scooped by
?
March 2, 2018 9:34 AM
|
We report the discovery that strains of Staphylococcus epidermidis produce 6- N -hydroxyaminopurine (6-HAP), a molecule that inhibits DNA polymerase activity. In culture, 6-HAP selectively inhibited proliferation of tumor lines but did not inhibit primary keratinocytes. Resistance to 6-HAP was associated with the expression of mitochondrial amidoxime reducing components, enzymes that were not observed in cells sensitive to this compound. Intravenous injection of 6-HAP in mice suppressed the growth of B16F10 melanoma without evidence of systemic toxicity. Colonization of mice with an S. epidermidis strain producing 6-HAP reduced the incidence of ultraviolet-induced tumors compared to mice colonized by a control strain that did not produce 6-HAP. S. epidermidis strains producing 6-HAP were found in the metagenome from multiple healthy human subjects, suggesting that the microbiome of some individuals may confer protection against skin cancer. These findings show a new role for skin commensal bacteria in host defense.

|
Scooped by
?
March 2, 2018 8:36 AM
|
Nitrous oxide (N2O) is a greenhouse gas that is also capable of destroying the ozone layer1. Agricultural soil is the largest source of N2O (ref. 2). Soybean is a globally important leguminous crop, and hosts symbiotic nitrogen-fixing soil bacteria (rhizobia) that can also produce N2O (ref. 3). In agricultural soil, N2O is emitted from fertilizer and soil nitrogen. In soybean ecosystems, N2O is also emitted from the degradation of the root nodules4. Organic nitrogen inside the nodules is mineralized to NH4+, followed by nitrification and denitrification that produce N2O. N2O is then emitted into the atmosphere or is further reduced to N2 by N2O reductase (N2OR), which is encoded by the nosZ gene. Pure culture and vermiculite pot experiments showed lower N2O emission by nosZ+ strains5 and nosZ++ strains (mutants with increased N2OR activity)6 of Bradyrhizobium japonicum than by nosZ− strains. A pot experiment using soil confirmed these results7. Although enhancing N2OR activity has been suggested as a N2O mitigation option8,9, this has never been tested in the field. Here, we show that post-harvest N2O emission from soybean ecosystems due to degradation of nodules can be mitigated by inoculation of nosZ+ and non-genetically modified organism nosZ++ strains of B. japonicum at a field scale.

|
Scooped by
?
March 1, 2018 11:52 PM
|
Microbes inhabiting the gut play an essential role in our biology, but could these gastrointestinal dwellers also shape our evolution?

|
Scooped by
?
March 1, 2018 8:35 AM
|
The soil bacterium Pseudomonas putida is rapidly becoming a platform of choice for applications that require a microbial host highly resistant to different types of stresses and elevated rates of reducing power regeneration. P. putida is capable of growing in a wide variety of carbon sources that range from simple sugars to complex substrates such as aromatic compounds. Interestingly, the growth of the reference strain KT2440 on glycerol as the sole carbon source is characterized by a prolonged lag phase, not observed with other carbon substrates. This macroscopic phenomenon has been shown to be connected with the stochastic expression of the glp genes, which encode the enzymes needed for glycerol processing. In this protocol, we propose a general procedure to examine bacterial growth in small-scale cultures while monitoring the metabolic activity of individual cells. Assessing the metabolic capacity of single bacteria by means of fluorescence microscopy and flow cytometry, in combination with the analysis of the temporal takeoff of growth in single-cell cultures, is a simple and easy-to-implement approach. It can help to understand the link between macroscopic phenotypes (e.g., microbial growth in batch cultures) and stochastic phenomena at the genetic level. The implementation of these methodologies revealed that the adoption of a glycerol-metabolizing regime by P. putida KT2440 is not the result of a gradual change in the whole population, but it rather reflects a time-dependent bimodal switch between metabolically inactive (i.e., not growing) to fully active (i.e., growing) bacteria.
|
 Your new post is loading...
Your new post is loading...

















rubio