 Your new post is loading...

|
Scooped by
?
April 30, 2021 1:24 AM
|
Human health is dependent on a plentiful and nutritious supply of food, primarily derived from crop plants. Rhythmic supply of light as a result of the day and night cycle led to the evolution of circadian clocks that modulate most plant physiology, photosynthesis, metabolism, and development. To regulate crop traits and adaptation, breeders have indirectly selected for variation at circadian genes. The pervasive impact of the circadian system on crops suggests that future food production might be improved by modifying circadian rhythms, engineering the timing of transgene expression, and applying agricultural treatments at the most effective time of day. We describe the applied research required to take advantage of circadian biology in agriculture to increase production and reduce inputs.

|
Scooped by
?
April 30, 2021 1:00 AM
|
CO2 sequestration engineering is an attractive strategy for achieving carbon- and energy-efficient bioproduction. However, the efficiency of heterotrophic CO2 sequestration is limited by bioproduct dependence and energy deficiency. Here, modular CO2 sequestration engineering was developed to produce target chemicals by integrating synthetic CO2 fixation and CO2 mitigation modules. A synthetic CO2 fixation pathway was designed, and then enhanced by light-driven reducing power using self-assembled cadmium sulfide nanoparticles. Next, a CO2 mitigation switch was designed, and then optimized by light-driven energy via proteorhodopsin. Finally, by integrating CO2 fixation and CO2 mitigation modules, the efficiency of CO2 sequestration was notably enhanced in Escherichia coli and the yields of l-malate and butyrate were increased to 1.48 and 0.79 mol/mol glucose, respectively, reaching theoretical yields. This CO2 sequestration system provides an efficient platform for channelling CO2 into value-added chemicals.

|
Scooped by
?
April 29, 2021 11:24 PM
|
Microorganisms play vital roles in modulating organic matter decomposition and nutrient cycling in soil ecosystems. The enzyme latch paradigm posits microbial degradation of polyphenols is hindered in anoxic peat leading to polyphenol accumulation, and consequently diminished microbial activity. This model assumes that polyphenols are microbially unavailable under anoxia, a supposition that has not been thoroughly investigated in any soil type. Here, we use anoxic soil reactors amended with and without a chemically defined polyphenol to test this hypothesis, employing metabolomics and genome-resolved metaproteomics to interrogate soil microbial polyphenol metabolism. Challenging the idea that polyphenols are not bioavailable under anoxia, we provide metabolite evidence that polyphenols are depolymerized, resulting in monomer accumulation, followed by the generation of small phenolic degradation products. Further, we show that soil microbiome function is maintained, and possibly enhanced, with polyphenol addition. In summary, this study provides chemical and enzymatic evidence that some soil microbiota can degrade polyphenols under anoxia and subvert the assumed polyphenol lock on soil microbial metabolism.

|
Scooped by
?
April 29, 2021 7:04 PM
|
To sustain growth in fluctuating environments microbial organisms must respond appropriately. The response generally requires the synthesis of novel proteins, but this synthesis can be impeded due to the depletion of biosynthetic precursors when growth conditions vary. Microbes must thus devise effective response strategies to manage depleting precursors. To better understand these strategies, we here investigate the active response of Escherichia coli to changes in nutrient conditions, connecting transient gene-expression behavior to growth phenotypes. By synthetically modifying the gene expression during changing growth conditions, we show how the competition by genes for the limited protein synthesis capacity constrains the cellular response. Despite this constraint, cells substantially express genes that are not required, severely slowing down the response. These findings highlight that cells do not optimize growth and recovery in every encountered environment but rather exhibit hardwired response strategies that may have evolved to promote growth in their native environment and include the regulation of multiple genes. The constraint and the suboptimality of the cellular response uncovered in this study provides a conceptual framework relevant for many research applications, from the prediction of evolution and adaptation to the improvement of gene circuits in biotechnology.

|
Scooped by
?
April 29, 2021 12:51 AM
|
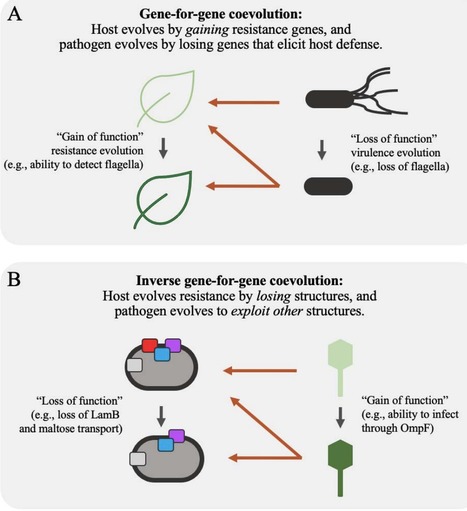
Bacteria often evolve resistance to phage through the loss or modification of cell-surface receptors. In Escherichia coli and phage λ, such resistance can catalyze a coevolutionary arms race focused on host and phage structures that interact at the outer membrane. Here, we analyze another facet of this arms race involving interactions at the inner membrane, whereby E. coli evolves mutations in mannose permease-encoding genes manY and manZ that impair λ′s ability to eject its DNA into the cytoplasm. We show that these man mutants arose concurrently with the arms race at the outer membrane. We tested the hypothesis that λ evolved an additional counter-defense that allowed them to infect bacteria with deleted man genes. The deletions severely impaired the ancestral λ, but some evolved phage grew well on the deletion mutants, indicating they regained infectivity by evolving the ability to infect hosts independently of the mannose permease. This coevolutionary arms race fulfills the model of an inverse-gene-for-gene infection network. Taken together, the interactions at both the outer and inner membranes reveal that coevolutionary arms races can be richer and more complex than is often appreciated.

|
Scooped by
?
April 29, 2021 12:33 AM
|
Bacterial biofilms demonstrate high broad-spectrum adaptive antibiotic resistance and cause two thirds of all infections, but there is a lack of approved antibiofilm agents. Unlike the standard minimal inhibitory concentration assay to assess antibacterial activity against planktonic cells, there is no standardized method to evaluate biofilm inhibition and/or eradication capacity of novel antibiofilm compounds. The protocol described here outlines simple and reproducible methods for assessing the biofilm inhibition and eradication capacities of novel antibiofilm agents against adherent bacterial biofilms grown in 96-well microtiter plates. It employs two inexpensive dyes: crystal violet to stain adhered biofilm biomass and 2,3,5-triphenyl tetrazolium chloride to quantify metabolism of the biofilm cells. The procedure is accessible to any laboratory with a plate reader, requires minimal technical expertise or training and takes 4 or 5 d to complete. Recommendations for how biofilm inhibition and eradication results should be interpreted and presented are also described. This protocol outlines simple and reproducible methods for assessing the biofilm inhibition and eradication capacities of novel antibiofilm agents against adherent bacterial biofilms grown in 96-well microtiter plates.

|
Scooped by
?
April 28, 2021 11:56 PM
|
The early detection of blood in urine (hematuria) can play a crucial role in the treatment of serious diseases (e.g., infections, kidney disease, schistosomiasis, and cancer). Therefore, the development of low-cost portable biosensors for blood detection in urine has become necessary. Here, we designed an ultrasensitive whole-cell bacterial biosensor interfaced with an optoelectronic measurement module for heme detection in urine. Heme is a red blood cells (RBCs) component that is liberated from lysed cells. The bacterial biosensor includes Escherichia coli cells carrying a heme-sensitive synthetic promoter integrated with a luciferase reporter (luxCDABE) from Photorhabdus luminescens. To improve the bacterial biosensor performance, we re-engineered the genetic structure of luxCDABE operon by splitting it into two parts (luxCDE and luxAB). The luxCDE genes were regulated by the heme-sensitive promoter, and the luxAB genes were regulated by either constitutive or inducible promoters. We examined the genetic circuit’s performance in synthetic urine diluent supplied with heme and in human urine supplied with lysed blood. Finally, we interfaced the bacterial biosensor with a light detection setup based on a commercial optical measurement single-photon avalanche photodiode (SPAD). The whole-cell biosensor was tested in human urine with lysed blood, demonstrating a low-cost, portable, and easy-to-use hematuria detection with an ON-to-OFF ratio of 6.5-fold for blood levels from 5 × 104 to 5 × 105 RBC per mL of human urine.

|
Scooped by
?
April 28, 2021 11:29 PM
|
Bacterial behavior is the outcome of both molecular mechanisms within each cell and interactions between cells in the context of their environment. Whereas whole-cell models simulate a single cell's behavior using molecular mechanisms, agent-based models simulate many agents independently acting and interacting to generate complex collective phenomena. To synthesize agent-based and whole-cell modeling, we used a novel model integration software, called Vivarium, to construct an agent-based model of E. coli colonies where each agent is represented by a current source code snapshot from the E. coli Whole-Cell Modeling Project and interacts with other cells in a shared spatial environment. The result is the first "whole-colony" computational model that mechanistically links expression of individual proteins to a population-level phenotype. Simulated colonies exhibit heterogeneous effects on their environments, heterogeneous gene expression, and media-dependent growth. Extending the cellular model with mechanisms of antibiotic susceptibility and resistance, our model also suggested that variation in the expression level of the beta-lactamase AmpC, and not of the multi-drug efflux pump AcrAB-TolC, was the key mechanistic driver of survival in the presence of nitrocefin. We see this as a significant step forward in the creation of more comprehensive multi-scale models, and it broadens the range of phenomena that can be modeled in mechanistic terms.

|
Scooped by
?
April 28, 2021 6:03 PM
|
A major outstanding challenge in the fields of biological research, synthetic biology and cell-based medicine is the difficulty of visualizing the function of natural and engineered cells noninvasively inside opaque organisms. Ultrasound imaging has the potential to address this challenge as a widely available technique with a tissue penetration of several centimeters and a spatial resolution below 100 um. Recently, the first genetically encoded reporter molecules were developed based on bacterial gas vesicles to link ultrasound signals to molecular and cellular function. However, the properties of these first-generation acoustic reporter genes (ARGs) resulted in limited sensitivity and specificity for imaging in the in vivo context. Here, we describe second-generation ARGs with greatly improved acoustic properties and expression characteristics. We identified these ARGs through a systematic phylogenetic screen of candidate gas vesicle gene clusters from diverse bacteria and archaea. The resulting constructs offer major qualitative and quantitative improvements, including the ability to produce nonlinear ultrasound contrast to distinguish their signals from those of background tissues, and a reduced burden of expression in probiotic hosts. We demonstrate the utility of these next-generation ARGs by imaging the in situ gene expression of tumor-homing probiotic bacteria, revealing the unique spatial distribution of tumor colonization by these cells noninvasively in living subjects.

|
Scooped by
?
April 28, 2021 3:18 AM
|
Like other plant compartments, the seed harbors a microbiome. The members of the seed microbiome are the first to colonize a germinating seedling, and they initiate the trajectory of microbiome assembly for the next plant generation. Therefore, the members of the seed microbiome are important for the dynamics of plant microbiome assembly and the vertical transmission of potentially beneficial symbionts. However, it remains challenging to assess the microbiome at the individual seed level (and, therefore, for the future individual plant) due to low endophytic microbial biomass, seed exudates that can select for particular members, and high plant and plastid contamination of resulting reads. Here, we report a protocol for extracting metagenomic DNA from an individual seed (common bean, Phaseolus vulgaris L.) with minimal disruption of host tissue, which we expect to be generalizable to other medium- and large- seed plant species. We applied this protocol to quantify the 16S rRNA V4 and ITS2 amplicon composition and variability for individual seeds harvested from replicate common bean plants grown under standard, controlled conditions to maintain health. Using metagenomic DNA extractions from individual seeds, we compared seed-to-seed, pod-to-pod, and plant-to-plant microbiomes, and found highest microbiome variability at the plant level. This suggests that several seeds from the same plant could be pooled for microbiome assessment, given experimental designs that apply treatments at the maternal plant level. This study adds protocols and insights to the growing toolkit of approaches to understand the plant-microbiome engagements that support the health of agricultural and environmental ecosystems.

|
Scooped by
?
April 28, 2021 2:04 AM
|
Bacteria have evolved small RNAs (sRNAs) to regulate numerous biological processes and stress responses. While sRNAs generally are considered to be "noncoding", a few have been found to also encode a small protein. Here we describe one such dual-function RNA that modulates carbon utilization in Escherichia coli. The 164 nucleotide RNA was previously shown to encode a 28 amino acid protein (denoted AzuC). We discovered the membrane-associated AzuC protein interacts with GlpD, the aerobic glycerol-3-phosphate dehydrogenase, leading to increased GlpD activity. Overexpression of the RNA encoding AzuC results in a growth defect in glycerol and galactose medium. The defect in galactose medium was still observed for a stop codon mutant derivative, suggesting a potential regulatory role for the RNA. Consistent with this observation, we found that cadA and galE are repressed by base pairing with the RNA (denoted AzuCR). Interestingly, translation of AzuC interferes with the observed repression of cadA and galE by AzuCR and base pairing interferes with AzuC translation, demonstrating that the translation and base pairing functions compete.

|
Scooped by
?
April 28, 2021 12:36 AM
|
Unlike nucleobase modifications in canonical restriction-modification systems, DNA phosphorothioate (PT) epigenetic modification occurs in the DNA sugar-phosphate backbone when the nonbridging oxygen is replaced by sulfur in a double-stranded (ds) or single-stranded (ss) manner governed by DndABCDE or SspABCD, respectively. SspABCD coupled with SspE constitutes a defense barrier in which SspE depends on sequence-specific PT modifications to exert its antiphage activity. Here, we identified a new type of ssDNA PT-based SspABCD-SspFGH defense system capable of providing protection against phages through a mode of action different from that of SspABCD-SspE. We provide further evidence that SspFGH damages non-PT-modified DNA and exerts antiphage activity by suppressing phage DNA replication. Despite their different defense mechanisms, SspFGH and SspE are compatible and pair simultaneously with one SspABCD module, greatly enhancing the protection against phages. Together with the observation that the sspBCD-sspFGH cassette is widely distributed in bacterial genomes, this study highlights the diversity of PT-based defense barriers and expands our knowledge of the arsenal of phage defense mechanisms.

|
Scooped by
?
April 28, 2021 12:14 AM
|
Vibrio cholerae causes the gastrointestinal illness cholera, which spreads throughout the globe in large pandemics. The current pandemic is caused by O1 El Tor biotype strains, whereas previous pandemics were caused by O1 classical biotype strains. El Tor V. cholerae is noted for its ability to acquire exogenous DNA through chitin-induced natural transformation, which has been exploited for genetic manipulation of El Tor strains in the laboratory. In contrast, the prototypical classical strain O395 lacks this ability, which was suspected to be due to a mutation in the regulatory gene hapR. HapR and the regulator TfoX control expression of a third competence regulator, QstR. We found that artificial induction of both TfoX and QstR in the presence of HapR in O395 was required for efficient DNA uptake. However, natural transformation in the classical strain is still orders of magnitude below that of an El Tor strain. O395 expressing HapR could also undergo natural transformation after growth on chitin, which could be increased by artificial induction of TfoX and/or QstR. A plasmid that expresses both TfoX and QstR was created that allowed for consistent DNA uptake in O395 carrying a hapR plasmid. This technique was also used to facilitate cotransformation into O395 of unmarked DNA (ΔlacZ, ΔflaA, ΔflgG) for multiplex genome editing by natural transformation (MuGENT). These results demonstrate that the classical biotype O395 strain is functionally capable of DNA uptake, which allows for the rapid genetic manipulation of its genome.
|

|
Scooped by
?
April 30, 2021 1:16 AM
|
Genomic DNA is composed of four standard nucleotides, each with a different nucleobase: adenine (A), thymine (T), cytosine (C), and guanine (G). These nucleobases form the genetic alphabet, ATCG, which is conserved across all domains of life. However, in 1977, the DNA virus cyanophage S-2L was discovered with all instances of A substituted with 2-aminoadenine (Z) throughout its genome (1, 2), forming the genetic alphabet ZTCG. Studies revealed interesting properties of Z-substituted DNA (dZ-DNA) (3–6), but little of Z synthesis was understood. On pages 516 and 512 of this issue, Sleiman et al. (7) and Zhou et al. (8), respectively, characterize viral Z biosynthesis. On page 520, Pezo et al. (9) identify a Z-specific DNA polymerase that is responsible for assembling dZ-DNA from nucleotides. All three studies identify additional “Z-genomes” in diverse bacteriophages (viruses that infect bacteria), which may have offered evolutionary advantages alongside standard ATCG DNA since life began.

|
Scooped by
?
April 29, 2021 11:26 PM
|
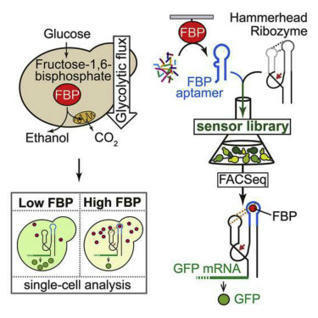
RNA-based sensors for intracellular metabolites are a promising solution to the emerging issue of metabolic heterogeneity. However, their development, i.e., the conversion of an aptamer into an in vivo-functional intracellular metabolite sensor, still harbors challenges. Here, we accomplished this for the glycolytic flux-signaling metabolite, fructose-1,6-bisphosphate (FBP). Starting from in vitro selection of an aptamer, we constructed device libraries with a hammerhead ribozyme as actuator. Using high-throughput screening in yeast with fluorescence-activated cell sorting (FACS), next-generation sequencing, and genetic-environmental perturbations to modulate the intracellular FBP levels, we identified a sensor that generates ratiometric fluorescent readout. An abrogated response in sensor mutants and occurrence of two sensor conformations—revealed by RNA structural probing—indicated in vivo riboswitching activity. Microscopy showed that the sensor can differentiate cells with different glycolytic fluxes within yeast populations, opening research avenues into metabolic heterogeneity. We demonstrate the possibility to generate RNA-based sensors for intracellular metabolites for which no natural metabolite-binding RNA element exits.

|
Scooped by
?
April 29, 2021 10:42 PM
|
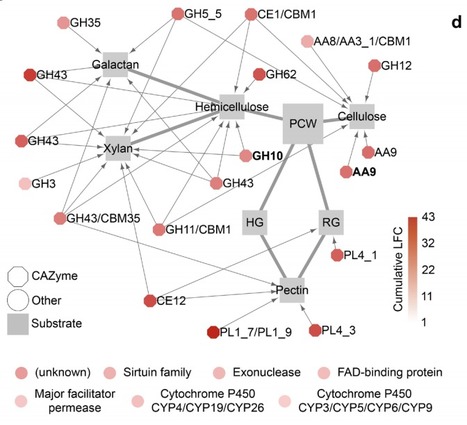
Roots of Arabidopsis thaliana do not engage in symbiotic associations with mycorrhizal fungi but host taxonomically diverse fungal communities that influence health and disease states. We sequenced the genomes of 41 fungal isolates representative of the A. thaliana root mycobiota for comparative analysis with 79 other plant-associated fungi. We report that root mycobiota members evolved from ancestors with diverse lifestyles and retained large repertoires of plant cell wall-degrading enzymes (PCWDEs) and effector-like small secreted proteins. We identified a set of 84 gene families predicting best endophytism, including families encoding PCWDEs acting on xylan (GH10) and cellulose (AA9). These genes also belong to a core transcriptional response induced by phylogenetically-distant mycobiota members in A. thaliana roots. Recolonization experiments with individual fungi indicated that strains with detrimental effects in mono-association with the host not only colonize roots more aggressively than those with beneficial activities but also dominate in natural root samples. We identified and validated the pectin degrading enzyme family PL1_7 as a key component linking aggressiveness of endophytic colonization to plant health.

|
Scooped by
?
April 29, 2021 2:49 AM
|
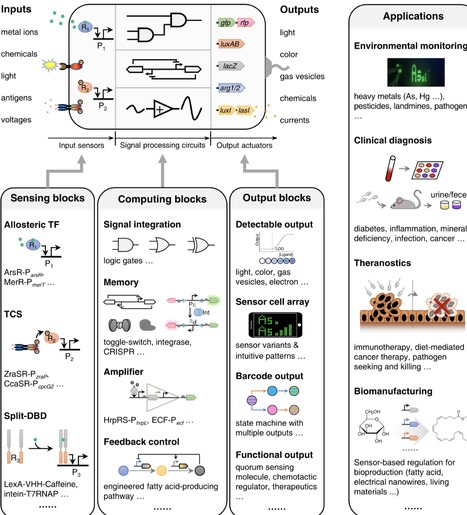
Synthetic biology offers new tools and capabilities of engineering cells with desired functions for example as new biosensing platforms leveraging engineered microbes. In the last two decades, bacterial cells have been programmed to sense and respond to various input cues for versatile purposes including environmental monitoring, disease diagnosis and adaptive biomanufacturing. Despite demonstrated proof-of-concept success in the lab, the real-world applications of microbial sensors have been restricted due to certain technical and societal limitations. Yet, most limitations can be addressed by new technological developments in synthetic biology such as circuit design, biocontainment and machine learning. Here, we summarize the latest advances in synthetic biology and discuss how they could accelerate the development, enhance the performance and address the present limitations of microbial sensors to facilitate their use in the field. We view that programmable living sensors are promising sensing platforms to achieve sustainable, affordable, and easy-to-use on-site detection in diverse settings.

|
Scooped by
?
April 29, 2021 12:42 AM
|
Promoters play a key role in influencing transcriptional regulation for fine-tuning expression of genes. Heterologous promoter engineering has been a widely used concept to control the level of transcription in all model organisms. The strength of a promoter is mainly determined by its nucleotide composition. Many promoter libraries have been curated but few have attempted to develop theoretical methods to predict the strength of promoters from its nucleotide sequence. Such theoretical methods are not only valuable in the design of promoters with specified strength, but are also meaningful to understand the mechanism of promoters in gene transcription. In this study, we present a theoretical model to describe the relationship between promoter strength and nucleotide sequence in Saccharomyces cerevisiae. We infer from our analysis that the -49 to 10 sequence with respect to the Transcription Start Site represents the minimal region that can be used to predict the promoter strength. We present an online tool https://qpromoters.com/ that takes advantage of this fact to quickly quantify the strength of the promoters.

|
Scooped by
?
April 29, 2021 12:13 AM
|
Despite numerous viral outbreaks in the last decade, including a devastating global pandemic, diagnostic and therapeutic technologies remain severely lacking. CRISPR-Cas systems have the potential to address these critical needs in the response against infectious disease. Initially discovered as the bacterial adaptive immune system, these systems provide a unique opportunity to create programmable, sequence-specific technologies for detection of viral nucleic acids and inhibition of viral replication. This review summarizes how CRISPR-Cas systems—in particular the recently discovered DNA-targeting Cas12 and RNA-targeting Cas13, both possessing a unique trans-cleavage activity—are being harnessed for viral diagnostics and therapies. We further highlight the numerous technologies whose development has accelerated in response to the COVID-19 pandemic.

|
Scooped by
?
April 28, 2021 11:43 PM
|
Barley has abundant anthocyanin-rich accessions, which renders it an ideal model to investigate the regulatory mechanism of anthocyanin biosynthesis. This study functionally characterized two transcription factors: Ant1 and Ant2. Sequence alignment showed that the coding sequences of Ant1 and Ant2 are conserved among 11 colored hulless barley and noncolored barley varieties. The expression profiles of Ant1 and Ant2 were divergent between species, and significantly higher expression was found in two colored Qingke accessions. The co-expression of Ant1 and Ant2 resulted in purple pigmentation in transient transformation systems via the promotion of the transcription of four structural genes. Ant1 interacted with Ant2, and overexpression of Ant1 activated the transcription of Ant2. Moreover, overexpression of Ant1 led to anthocyanin accumulation in the pericarp and aleurone layer of transgenic barley grains. Overall, our results suggest that anthocyanin-enriched barley grains can be produced by manipulating Ant1 expression.

|
Scooped by
?
April 28, 2021 11:19 PM
|
CRISPR-Cas systems recognize foreign genetic material using CRISPR RNAs (crRNAs). In Type II systems, a trans-activating crRNA (tracrRNA) hybridizes to crRNAs to drive their processing and utilization by Cas9. While analyzing Cas9-RNA complexes from Campylobacter jejuni, we discovered tracrRNA hybridizing to cellular RNAs, leading to formation of “noncanonical” crRNAs capable of guiding DNA targeting by Cas9. Our discovery inspired the engineering of reprogrammed tracrRNAs that link the presence of any RNA-of-interest to DNA targeting with different Cas9 orthologs. This capability became the basis for a multiplexable diagnostic platform termed LEOPARD (Leveraging Engineered tracrRNAs and On-target DNAs for PArallel RNA Detection). LEOPARD allowed simultaneous detection of RNAs from different viruses in one test and distinguished SARS-CoV-2 and its D614G variant with single-base resolution in patient samples.

|
Scooped by
?
April 28, 2021 5:39 PM
|
Isolating single phages using plaque assays is a laborious and time-consuming process. Whether single isolated phages are the most lyse-effective, the most abundant in viromes, or the ones with highest ability to plaque on solid media is not well known. With the increasing accessibility of high-throughput sequencing, metaviromics is often used to describe viruses in environmental samples. By extracting and sequencing metaviromes from organic waste with and without exposure to a host-of-interest, we show a host-related phage community's shift, as well as identify the most enriched phages. Moreover, we isolated plaque-forming single phages using the same virome-host matrix to observe how enrichments in liquid media corresponds to the metaviromic data. In this study, we observed a significant shift (p = 0.015) of the 47 identified putative Pseudomonas phages with a minimum 2-fold change above 0 in read abundance when adding a Pseudomonas syringae DC3000 host. Surprisingly, it appears that only two out of five plaque-forming phages from the same organic waste sample, targeting the Pseudomonas strain, was highly abundant in the metavirome, while the other three were almost absent despite host exposure. Lastly, our sequencing results highlights how long reads from Oxford Nanopore elevates the assembly quality of metaviromes, compared to short reads alone.

|
Scooped by
?
April 28, 2021 2:54 AM
|
In bacterial synthetic biology, whole genome transplantation has been achieved only in mycoplasmas that contain a small genome and are competent for foreign genome uptake. In this study, we developed Escherichia coli strains programmed by three 1-megabase (Mb) chromosomes by splitting the 3-Mb chromosome of a genome-reduced strain. The first split-chromosome retains the original replication origin (oriC) and partitioning (par) system. The second one has an oriC and the par locus from the F plasmid, while the third one has the ori and par locus of the Vibrio tubiashii secondary chromosome. The tripartite-genome cells maintained the rod-shaped form and grew only twice as slowly as their parent, allowing their further genetic engineering. A proportion of these 1-Mb chromosomes were purified as covalently closed supercoiled molecules with a conventional alkaline lysis method and anion exchange columns. Furthermore, the second and third chromosomes could be individually electroporated into competent cells. In contrast, the first split-chromosome was not able to coexist with another chromosome carrying the same origin region. However, it was exchangeable via conjugation between tripartite-genome strains by using different selection markers. We believe that this E. coli-based technology has the potential to greatly accelerate synthetic biology and synthetic genomics.

|
Scooped by
?
April 28, 2021 1:26 AM
|
The cell cycle is the process by which eukaryotic cells replicate. Yeast cells cycle asynchronously with each cell in the population budding at a different time. Although there are several experimental approaches to synchronize cells, these usually work only in the short-term. Here, we build a cyber-genetic system to achieve long-term synchronisation of the cell population, by interfacing genetically modified yeast cells with a computer by means of microfluidics to dynamically change medium, and a microscope to estimate cell cycle phases of individual cells. The computer implements a controller algorithm to decide when, and for how long, to change the growth medium to synchronise the cell-cycle across the population. Our work builds upon solid theoretical foundations provided by Control Engineering. In addition to providing an avenue for yeast cell cycle synchronisation, our work shows that control engineering can be used to automatically steer complex biological processes towards desired behaviours similarly to what is currently done with robots and autonomous vehicles.

|
Scooped by
?
April 28, 2021 12:24 AM
|
Extracellular electron transfer (EET) is an important biological process in microbial physiology as found in dissimilatory metal oxidation/reduction and interspecies electron transfer in syntrophy in natural environments. EET also plays a critical role in microorganisms relevant to environmental biotechnology in metal-contaminated areas, metal corrosion, bioelectrochemical systems, and anaerobic digesters. Geobacter species exist in a diversity of natural and artificial environments. One of the outstanding features of Geobacter species is the capability of direct EET with solid electron donors and acceptors, including metals, electrodes, and other cells. Therefore, Geobacter species are pivotal in environmental biogeochemical cycles and biotechnology applications. Geobacter sulfurreducens, a representative Geobacter species, has been studied for direct EET as a model microorganism. G. sulfurreducens employs electrically conductive pili (e-pili) and c-type cytochromes for the direct EET. The biological function and electronics applications of the e-pili have been reviewed recently, and this review focuses on the cytochromes. Geobacter species have an unusually large number of cytochromes encoded in their genomes. Unlike most other microorganisms, Geobacter species localize multiple cytochromes in each subcellular fraction, outer membrane, periplasm, and inner membrane, as well as in the extracellular space, and differentially utilize these cytochromes for EET with various electron donors and acceptors. Some of the cytochromes are functionally redundant. Thus, the EET in Geobacter is complicated. Geobacter coordinates the cytochromes with other cellular components in the elaborate EET system to flourish in the environment.
|
 Your new post is loading...
Your new post is loading...




























review