 Your new post is loading...

|
Scooped by
?
April 28, 2021 12:14 AM
|
Vibrio cholerae causes the gastrointestinal illness cholera, which spreads throughout the globe in large pandemics. The current pandemic is caused by O1 El Tor biotype strains, whereas previous pandemics were caused by O1 classical biotype strains. El Tor V. cholerae is noted for its ability to acquire exogenous DNA through chitin-induced natural transformation, which has been exploited for genetic manipulation of El Tor strains in the laboratory. In contrast, the prototypical classical strain O395 lacks this ability, which was suspected to be due to a mutation in the regulatory gene hapR. HapR and the regulator TfoX control expression of a third competence regulator, QstR. We found that artificial induction of both TfoX and QstR in the presence of HapR in O395 was required for efficient DNA uptake. However, natural transformation in the classical strain is still orders of magnitude below that of an El Tor strain. O395 expressing HapR could also undergo natural transformation after growth on chitin, which could be increased by artificial induction of TfoX and/or QstR. A plasmid that expresses both TfoX and QstR was created that allowed for consistent DNA uptake in O395 carrying a hapR plasmid. This technique was also used to facilitate cotransformation into O395 of unmarked DNA (ΔlacZ, ΔflaA, ΔflgG) for multiplex genome editing by natural transformation (MuGENT). These results demonstrate that the classical biotype O395 strain is functionally capable of DNA uptake, which allows for the rapid genetic manipulation of its genome.

|
Scooped by
?
April 27, 2021 8:00 PM
|
Technological advances in rare DNA mutations detection have revolutionized the diagnosis and monitoring of tumors, but they are still limited by the lack of supersensitive and high-coverage procedures for identifying low-abundance mutations. Here, we describe a single-tube, multiplex PCR-based system, A-Star, that involves a hyperthermophilic Argonaute from Pyrococcus furiosus (PfAgo) for highly efficient detection of rare mutations beneficial from its compatibility with DNA polymerase. This novel technique uses a specific guide design strategy to allow PfAgo selective cleavage with single-nucleotide resolution at 94°C, thus mostly eliminating wild-type DNA in the denaturation step and efficiently amplifying rare mutant DNA during the PCR process. The integrated single-tube system achieved great efficiency for enriching rare mutations compared with a divided system separating the cleavage and amplification. Thus, A-Star enables easy detection and quantification of 0.01% rare mutations with ≥5500-fold increase in efficiency. The feasibility of A-Star was also demonstrated for detecting oncogenic mutations in solid tumor tissues and blood samples. Remarkably, A-Star achieved simultaneous detection of multiple oncogenes through a simple single-tube reaction by orthogonal guide-directed specific cleavage. This study demonstrates a supersensitive and rapid nucleic acid detection system with promising potential for both research and therapeutic applications.

|
Scooped by
?
April 27, 2021 6:28 PM
|
The advancements in genome editing techniques over the past years have rekindled interest in rational metabolic engineering strategies. While Metabolic Control Analysis (MCA) is a well-established method for quantifying the effects of metabolic engineering interventions on flows in metabolic networks and metabolite concentrations, it does not consider the physiological limitations of the cellular environment and metabolic engineering design constraints. We report here a constraint-based framework, Network Response Analysis (NRA), for rational genetic strain design. NRA is cast as a Mixed-Integer Linear Programming problem that integrates MCA, Thermodynamically-based Flux Analysis (TFA), biologically relevant constraints, as well as genome editing restrictions into a comprehensive platform for identifying metabolic engineering targets. We show that the NRA formulation and its core constraints are equivalent to the ones of Flux Balance Analysis (FBA) and TFA, which allows it to be used for a wide range of optimization criteria and with various physiological constraints. We also show how the parametrization and introduction of biological constraints enhance the NRA formulation compared to the classical MCA approach, and we demonstrate its features and ability to generate multiple alternative optimal strategies given several user-defined boundaries and objectives. In summary, NRA is a sophisticated alternative to classical MCA for rational metabolic engineering that accommodates the incorporation of physiological data at metabolic flux, metabolite concentration, and enzyme expression levels.

|
Scooped by
?
April 27, 2021 1:56 AM
|
Compartmentation of proteins into biomolecular condensates or membraneless organelles formed by phase separation is an emerging principle for the regulation of cellular processes. Creating synthetic condensates that accommodate specific intracellular proteins on demand would have various applications in chemical biology, cell engineering, and synthetic biology. Here, we report the construction of synthetic protein condensates capable of recruiting and/or releasing proteins of interest in living mammalian cells in response to a small molecule or light. By a modular combination of a tandem fusion of two oligomeric proteins, which forms phase-separated synthetic protein condensates in cells, with a chemically induced dimerization tool, we first created a chemogenetic protein condensate system that can rapidly recruit target proteins from the cytoplasm to the condensates by addition of a small-molecule dimerizer. We next coupled the protein-recruiting condensate system with an engineered proximity-dependent protease, which gave a second protein condensate system wherein target proteins previously expressed inside the condensates are released into the cytoplasm by small-molecule-triggered protease recruitment. Furthermore, an optogenetic condensate system that allows reversible release and sequestration of protein activity in a repeatable manner using light was constructed successfully. These condensate systems were applicable to control protein activity and cellular processes such as membrane ruffling and ERK signaling in a time scale of minutes. This proof-of-principle work provides a new platform for chemogenetic and optogenetic control of protein activity in mammalian cells and represents a step toward tailor-made engineering of synthetic protein condensate-based soft materials with various functionalities for biological and biomedical applications.

|
Scooped by
?
April 27, 2021 12:03 AM
|
Lignocellulose, the structural component of plant cells, is a major agricultural byproduct and the most abundant terrestrial source of biopolymers on Earth. The complex and insoluble nature of lignocellulose limits its conversion into value-added commodities, and currently, efficient transformation requires expensive pretreatments and high loadings of enzymes. Here, we report on a fungus from the Parascedosporium genus, isolated from a wheat-straw composting community, that secretes a large and diverse array of carbohydrate-active enzymes (CAZymes) when grown on lignocellulosic substrates. We describe an oxidase activity that cleaves the major β-ether units in lignin, thereby releasing the flavonoid tricin from monocot lignin and enhancing the digestion of lignocellulose by polysaccharidase mixtures. We show that the enzyme, which holds potential for the biorefining industry, is widely distributed among lignocellulose-degrading fungi from the Sordariomycetes phylum.

|
Scooped by
?
April 26, 2021 2:19 PM
|
We previously developed REXER (Replicon EXcision Enhanced Recombination); this method enables the replacement of >100 kb of the Escherichia coli genome with synthetic DNA in a single step and allows the rapid identification of non-viable or otherwise problematic sequences with nucleotide resolution. Iterative repetition of REXER (GENESIS, GENomE Stepwise Interchange Synthesis) enables stepwise replacement of longer contiguous sections of genomic DNA with synthetic DNA, and even the replacement of the entire E. coli genome with synthetic DNA. Here we detail protocols for REXER and GENESIS. A standard REXER protocol typically takes 7–10 days to complete. Our description encompasses (i) synthetic DNA design, (ii) assembly of synthetic DNA constructs, (iii) utilization of CRISPR–Cas9 coupled to lambda-red recombination and positive/negative selection to enable the high-fidelity replacement of genomic DNA with synthetic DNA (or insertion of synthetic DNA), (iv) evaluation of the success of the integration and replacement and (v) identification of non-tolerated synthetic DNA sequences with nucleotide resolution. This protocol provides a set of precise genome engineering methods to create custom synthetic E. coli genomes.

|
Scooped by
?
April 26, 2021 12:49 PM
|
The l-arabinose-responsive AraC and its cognate PBAD promoter underlie one of the most often used chemically inducible prokaryotic gene expression systems in microbiology and synthetic biology. Here, we change the sensing capability of AraC from l-arabinose to blue light, making its dimerization and the resulting PBAD activation light-inducible. We engineer an entire family of blue light-inducible AraC dimers in Escherichia coli (BLADE) to control gene expression in space and time. We show that BLADE can be used with pre-existing l-arabinose-responsive plasmids and strains, enabling optogenetic experiments without the need to clone. Furthermore, we apply BLADE to control, with light, the catabolism of l-arabinose, thus externally steering bacterial growth with a simple transformation step. Our work establishes BLADE as a highly practical and effective optogenetic tool with plug-and-play functionality—features that we hope will accelerate the broader adoption of optogenetics and the realization of its vast potential in microbiology, synthetic biology and biotechnology.

|
Scooped by
?
April 26, 2021 12:58 AM
|
Antibiotic resistance spreads among bacteria through horizontal transfer of antibiotic resistance genes (ARGs). Here, we set out to determine predictive features of ARG transfer among bacterial clades. We use a statistical framework to identify putative horizontally transferred ARGs and the groups of bacteria that disseminate them. We identify 152 gene exchange networks containing 22,963 bacterial genomes. Analysis of ARG-surrounding sequences identify genes encoding putative mobilization elements such as transposases and integrases that may be involved in gene transfer between genomes. Certain ARGs appear to be frequently mobilised by different mobile genetic elements. We characterise the phylogenetic reach of these mobilisation elements to predict the potential future dissemination of known ARGs. Using a separate database with 472,798 genomes from Streptococcaceae, Staphylococcaceae and Enterobacteriaceae, we confirm 34 of 94 predicted mobilisations. We explore transfer barriers beyond mobilisation and show experimentally that physiological constraints of the host can explain why specific genes are largely confined to Gram-negative bacteria although their mobile elements support dissemination to Gram-positive bacteria. Our approach may potentially enable better risk assessment of future resistance gene dissemination.

|
Scooped by
?
April 26, 2021 12:16 AM
|
The genus Pseudomonas can enhance plant resistance to a range of pathogens and herbivores. However, resistance to these different classes of plant antagonists is mediated by different molecular mechanisms, and the extent to which induced systemic resistance by Pseudomonas can simultaneously protect plants against both pathogens and herbivores remains unclear. We screened 12 root-colonizing Pseudomonas strains to assess their ability to induce resistance in Arabidopsis thaliana against a foliar pathogen (Pseudomonas syringae DC3000) and a chewing herbivore (Spodoptera littoralis). None of our 12 strains increased plant resistance against herbivory; however, four strains enhanced pathogen resistance, and one of these (Pseudomonas strain P97-38) also made plants more susceptible to herbivory. Phytohormone analyses revealed stronger SA induction in plants colonized by P97-38 (vs controls) following subsequent pathogen infection but weaker induction of JA-mediated defenses following herbivory. We found no effects of P97-38 inoculation on herbivore-relevant nutrients such as sugars and protein, suggesting that the observed enhancement of susceptibility to S. littoralis is due to effects on plant defense chemistry rather than nutrition. These findings suggest that Pseudomonas strains that enhance plant resistance to pathogens may have neutral or negative effects on resistance to herbivores and provide insight into potential mechanisms associated with effects on different classes of plant antagonists. Improved understanding of these effects has potentially important implications for the use of rhizobacteria inoculation in agriculture.

|
Scooped by
?
April 25, 2021 4:55 PM
|
Fumarate was previously known to serve as an anaerobic electron acceptor by E. coli when colonizing the mammalian intestine, but the source of that fumarate was elusive. In this issue, Unden and coworkers demonstrate that l‐aspartic acid is the source of fumarate that drives anaerobic respiration by colonized E. coli (Schubert et al., 2021). Moreover, Schubert et al., establish that E. coli is able to grow anaerobically by using aspartate as a sole source of nitrogen. These groundbreaking findings indicate that a single amino acid – aspartate – supports anaerobic respiration and acquisition of nitrogen by E. coli in the intestine.

|
Scooped by
?
April 25, 2021 4:38 PM
|
The current perspective presents an outlook on developing gut-like bioreactors with immobilized probiotic bacteria using cellulose hydrogels. The innovative concept of using hydrogels to simulate the human gut environment by generating and maintaining pH and oxygen gradients in the gut-like bioreactors is discussed. Fundamentally, this approach presents novel methods of production as well as delivery of multiple strains of probiotics using bioreactors. The relevant existing synthesis methods of cellulose hydrogels are discussed for producing porous hydrogels. Harvesting methods of multiple strains are discussed in the context of encapsulation of probiotic bacteria immobilized on cellulose hydrogels. Furthermore, we also discuss recent advances in using cellulose hydrogels for encapsulation of probiotic bacteria. This perspective also highlights the mechanism of probiotic protection by cellulose hydrogels. Such novel gut-like hydrogel bioreactors will have the potential to simulate the human gut ecosystem in the laboratory and stimulate new research on gut microbiota.

|
Scooped by
?
April 25, 2021 4:23 PM
|
Rice leaf blight is caused by the bacterium Xanthomonas oryzae pv. oryzae (Xoo). The upregulated by transcription activator-like 1 (UPT) effector box in the promoter region of the rice Xa13 gene plays a key role in Xoo pathogenicity. Mutation of a key bacterial protein-binding site in the UPT box of Xa13 to abolish PXO99-induced Xa13 expression is a way to improve rice resistance to bacteria. Highly efficient generation and selection of transgene-free edited plants are helpful to shorten and simplify the gene editing-based breeding process. Selective elimination of transgenic pollen of T0 plants can enrich the proportion of T1 transgene-free offspring, and expression of a color marker gene in seeds makes the selection of T2 plants very convenient and efficient. In this study, a genome editing and multiplexed selection system was used to generate bacterial leaf blight-resistant and transgene-free rice plants. We introduced site-specific mutations into the UPT box using CRISPR/Cas12a technology to hamper with transcription-activator-like effector (TAL) protein binding and gene activation and generated genome-edited rice with improved bacterial blight resistance. Transgenic pollen of T0 plants was eliminated by pollen-specific expression of the α-amylase gene Zmaa1, and the proportion of transgene-free plants increased from 25 to 50% among single T-DNA insertion events in the T1 generation. Transgenic seeds were visually identified and discarded by specific aleuronic expression of DsRed, which reduced the cost by 50% and led to up to 98.64% accuracy for the selection of transgene-free edited plants.

|
Scooped by
?
April 25, 2021 3:06 PM
|
- Cable bacteria are sulfide‐oxidizing, filamentous bacteria which reduce toxic sulfide levels, suppress methane emissions, and drive nutrient and carbon cycling in sediments. Recently, cable bacteria have been found associated with roots of aquatic plants and rice (Oryza sativa). However, the extent to which cable bacteria are associated with aquatic plants in nature remains unexplored.
- Using newly generated and public 16S rRNA gene sequence datasets combined with fluorescence in situ hybridization, we investigated the distribution of cable bacteria around the roots of aquatic plants, encompassing seagrass (including seagrass seedlings), rice, freshwater and saltmarsh plants.
- Diverse cable bacteria were found associated with roots of 16 out of 28 plant species and at 36 out of 55 investigated sites, across four continents. Plant associated cable bacteria were confirmed across a variety of ecosystems, including marine coastal environments, estuaries, freshwater streams, isolated pristine lakes and intensive agricultural systems. This pattern indicates that this plant‐microbe relationship is globally widespread and neither obligate nor species‐specific.
- The occurrence of cable bacteria in plant rhizospheres may be of general importance to vegetation vitality, primary productivity, coastal restoration practices and greenhouse gas balance of rice fields and wetlands.
|

|
Scooped by
?
April 27, 2021 8:07 PM
|
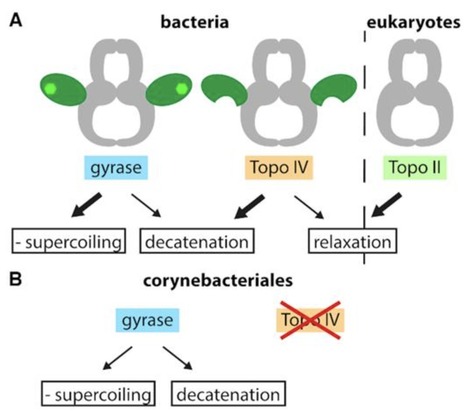
Type IIA topoisomerases catalyze a variety of different reactions: eukaryotic topoisomerase II relaxes DNA in an ATP-dependent reaction, whereas the bacterial representatives gyrase and topoisomerase IV (Topo IV) preferentially introduce negative supercoils into DNA (gyrase) or decatenate DNA (Topo IV). Gyrase and Topo IV perform separate, dedicated tasks during replication: gyrase removes positive supercoils in front, Topo IV removes pre-catenanes behind the replication fork. Despite their well-separated cellular functions, gyrase and Topo IV have an overlapping activity spectrum: gyrase is also able to catalyze DNA decatenation, although less efficiently than Topo IV. The balance between supercoiling and decatenation activities is different for gyrases from different organisms. Both enzymes consist of a conserved topoisomerase core and structurally divergent C-terminal domains (CTDs). Deletion of the entire CTD, mutation of a conserved motif and even by just a single point mutation within the CTD converts gyrase into a Topo IV-like enzyme, implicating the CTDs as the major determinant for function. Here, we summarize the structural and mechanistic features that make a type IIA topoisomerase a gyrase or a Topo IV, and discuss the implications for type IIA topoisomerase evolution.

|
Scooped by
?
April 27, 2021 7:10 PM
|
The isoelectric point is the pH at which a particular molecule is electrically neutral due to the equilibrium of positive and negative charges. In proteins and peptides, this depends on the dissociation constant (pKa) of charged groups of seven amino acids and NH+ and COO− groups at polypeptide termini. Information regarding isoelectric point and pKa is extensively used in two-dimensional gel electrophoresis (2D-PAGE), capillary isoelectric focusing (cIEF), crystallisation, and mass spectrometry. Therefore, there is a strong need for the in silico prediction of isoelectric point and pKa values. In this paper, I present Isoelectric Point Calculator 2.0 (IPC 2.0), a web server for the prediction of isoelectric points and pKa values using a mixture of deep learning and support vector regression models. The prediction accuracy (RMSD) of IPC 2.0 for proteins and peptides outperforms previous algorithms: 0.848 versus 0.868 and 0.222 versus 0.405, respectively. Moreover, the IPC 2.0 prediction of pKa using sequence information alone was better than the prediction from structure-based methods (0.576 versus 0.826) and a few folds faster. The IPC 2.0 webserver is freely available at www.ipc2-isoelectric-point.org

|
Scooped by
?
April 27, 2021 2:19 AM
|
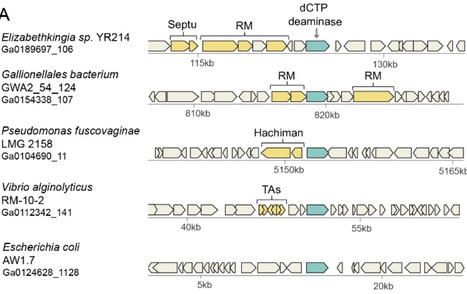
DNA viruses and retroviruses need to consume large quantities of deoxynucleotides (dNTPs) when replicating within infected cells. The human antiviral factor SAMHD1 takes advantage of this vulnerability in the viral life cycle, and inhibits viral replication by degrading dNTPs into their constituent deoxynucleosides and inorganic phosphate. In this study we report that bacteria employ a similar strategy to defend against phage infection. We found a family of defensive dCTP deaminase proteins that, in response to phage infection, convert dCTP into deoxy-uracil nucleotides. A second family of phage resistance genes encode dGTPase enzymes, which degrade dGTP into phosphate-free deoxy-guanosine (dG) and are distant homologs of the human SAMHD1. Our results show that the defensive proteins completely eliminate the specific deoxynucleotide (either dCTP or dGTP) from the nucleotide pool during phage infection, thus starving the phage of an essential DNA building block and halting its replication. Both defensive genes are found in a diverse set of bacterial species and are specifically enriched in Vibrio genomes. Our study demonstrates that manipulation of the deoxynucleotide pool is a potent antiviral strategy shared by both prokaryotes and eukaryotes.

|
Scooped by
?
April 27, 2021 1:12 AM
|
A diverse community of microorganisms inhabits various parts of a plant. Recent findings indicate that perturbations to the normal microbiota can be associated with positive and negative effects on plant health. In this review, we discuss these findings in the context of understanding how microbiota homeostasis is regulated in plants for promoting health and/or for preventing dysbiosis.

|
Scooped by
?
April 26, 2021 11:53 PM
|
Fluorescent reporters have revolutionized modern applications in the fields of molecular and synthetic biology, enabling applications ranging from education to point-of-care diagnostics. Past advancements in these fields have primarily focused on improving reaction conditions, the development of new applications, and the broad dissemination of these technologies. However, field and classroom-based applications have remained limited in part due to the nature of fluorescent signal detection, which often requires the use of costly lab equipment to observe and quantify fluorescence readouts. Users without access to laboratory equipment rely on qualitative assessments of fluorescence, a process that remains highly variable from user-to-user even within the same classroom. To overcome this challenge, we have developed a foldable illuminator and incubator device to support field-applications of synthetic biology-based biosensors for education and diagnostics. The Fold-Illuminator is an affordable, portable, and recyclable device that allows for the visible detection of fluorescent biomolecules. The Fold-Illuminator's design allows for assembly in under 10 min, a user can then utilize the optional heating element to incubate biochemical reactions and visualize fluorescence outputs in a defined and light-controlled environment. Interchangeable LED strips and light-filtering screens provide modularity to pair with the fluorescence wavelengths of interest. The user can then unfold the device for convenient storage, transport, or even recycling. The cost for the Fold-Illuminator is $5.58 USD and is compatible with an optional heating element for an additional $3.98 cost, with potential for further reductions in cost for larger quantities. Open-source templates for cutting device parts from paper stock are provided for both printing and cutting by hand; cutting can also be achieved with consumer-grade smart cutting machines such as the Cricut®. Combined with the broad applications of fluorescent reporters, the Fold-Illuminator has the potential to improve access to fluorescence visualization and quantification for new users as well as emerging field applications.

|
Scooped by
?
April 26, 2021 2:05 PM
|
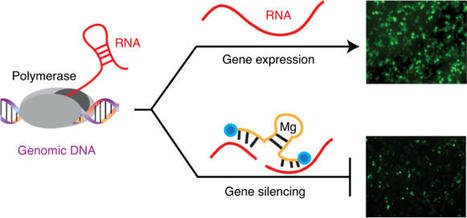
Efforts to use RNA-cleaving DNA enzymes (DNAzymes) as gene-silencing agents in therapeutic applications have stalled due to their low efficacy in clinical trials. Here we report a xeno-nucleic-acid-modified version of the classic DNAzyme 10–23 that achieves multiple-turnover activity under cellular conditions and resists nuclease digestion. The new reagent, X10–23, overcomes the problem of product inhibition, which limited previous 10–23 designs, using molecular chemotypes with DNA, 2′-fluoroarabino nucleic acid and α-l-threofuranosyl nucleic acid backbone architectures that balance the effects of enhanced biological stability with RNA hybridization and divalent metal ion coordination. In cultured mammalian cells, X10–23 facilitates persistent gene silencing by efficiently degrading exogenous and endogenous messenger RNA transcripts. Together, these results demonstrate that new molecular chemotypes can improve the activity and stability of DNAzymes, and may provide a new route for nucleic acid enzymes to reach the clinic.

|
Scooped by
?
April 26, 2021 1:09 AM
|
Agrobacterium tumefaciens, is a member of the Alphaproteobacteria that pathogenizes plants, and associates with biotic and abiotic surfaces via a single cellular pole. A. tumefaciens produces the unipolar polysaccharide (UPP) at the site of surface contact. UPP production is normally surface-contact inducible, but elevated levels of the second messenger cyclic diguanylate monophosphate (c-di-GMP) bypass this requirement. Multiple lines of evidence suggest that the UPP has a central polysaccharide component. Using an A. tumefaciens derivative with elevated cdGMP and mutationally disabled for other dispensable polysaccharides, a series of related genetic screens have identified a large number of genes involved in UPP biosynthesis, most of which are Wzx-Wzy-type polysaccharide biosynthetic components. Extensive analyses of UPP production in these mutants have revealed that the UPP is comprised of two genetically, chemically and spatially discrete forms of polysaccharide, and that each requires a specific Wzy-type polymerase. Other important biosynthetic, processing and regulatory functions for UPP production are also revealed, some of which are common to both polysaccharides, and a subset of which are specific to each species. Many of the UPP genes identified are conserved among diverse rhizobia, whereas others are more lineage specific.

|
Scooped by
?
April 26, 2021 12:27 AM
|
Biofilm formation is often attributed to post-harvest bacteria persistence on fresh produce and food handling surfaces. In this study, a predicted glycosyl hydrolase enzyme was expressed, purified and validated for removal of microbial biofilms from biotic and abiotic surfaces under conditions used for chemical cleaning agents. Crystal violet biofilm staining assays revealed that 0.1 mg/mL of enzyme inhibited up to 41% of biofilm formation by E. coli O157:H7, E. coli 25922, Salmonella Typhimurium, and Listeria monocytogenes. Further, the enzyme was effective at removing mature biofilms, providing a 35% improvement over rinsing with a saline solution alone. Additionally, a parallel-plate flow cell was used to directly observe and quantify the impact of enzyme rinses on E. coli O157:H7 cells adhered to spinach leaf surfaces. The presence of 1 mg/L enzyme resulted in nearly 6 times greater detachment rate coefficients than a DI water rinse, while the total cells removed from the surface increased from 10% to 25% over the 30 minute rinse time, reversing the initial phases of biofilm formation. Enzyme treatment of all 4 cell types resulted in significantly reduced cell surface hydrophobicity, and collapse of negatively stained E. coli 25922 cells imaged by electron microscopy, suggesting potential polysaccharide surface modification of enzyme-treated bacteria. Collectively, these results point to the broad substrate specificity and robustness of the enzyme to different types of biofilm stages, solution conditions and pathogen biofilm types, and may be useful as a method for removal or inhibition of bacterial biofilm formation.

|
Scooped by
?
April 25, 2021 4:58 PM
|
Transcription-factor-based biosensors (TFBs) are often used for metabolite detection, adaptive evolution, and metabolic flux control. However, designing TFBs with superior performance for applications in synthetic biology remains challenging. Specifically, natural TFBs often do not meet real-time detection requirements owing to their slow response times and inappropriate dynamic ranges, detection ranges, sensitivity, and selectivity. Furthermore, designing and optimizing complex dynamic regulation networks is time-consuming and labor-intensive. This Review highlights TFB-based applications and recent engineering strategies ranging from traditional trial-and-error approaches to novel computer-model-based rational design approaches. The limitations of the applications and these engineering strategies are additionally reviewed.

|
Scooped by
?
April 25, 2021 4:43 PM
|
Mycorrhizal symbiosis, comprising functionally distinctive plant‐fungus associations, mediates key plant population and community processes, and ultimately the functioning of terrestrial ecosystems. It is estimated that about 90% of the world’s vascular flora forms mycorrhizal symbioses with soil fungi. While this general estimate is probably adequate, there is a severe shortage of empirical information about mycorrhizal associations at the plant species level, with only around 5% of the world’s flora explored. Several database developments have emerged since the seminal work of Harley & Harley; extending both the number and the geography of plant species covered and defining and describing some key mycorrhizal traits of plant species – “mycorrhizal type” and “mycorrhizal status”. Nonetheless, this expansion poses new challenges connected with the compilation of global data based on heterogeneous sources with different practical and conceptual frameworks. Careful work developing consistent definitions and standardizing field and lab protocols is essential for harmonizing database content and avoiding critical inconsistencies.

|
Scooped by
?
April 25, 2021 4:28 PM
|
The chemical synthesis of monoatomic metallic copper is unfavorable and requires inert or reductive conditions and the use of toxic reagents. Here, we report the environmental extraction and conversion of CuSO4 ions into single-atom zero-valent copper (Cu0) by a copper-resistant bacterium isolated from a copper mine in Brazil. Furthermore, the biosynthetic mechanism of Cu0 production is proposed via proteomics analysis. This microbial conversion is carried out naturally under aerobic conditions eliminating toxic solvents. One of the most advanced commercially available transmission electron microscopy systems on the market (NeoArm) was used to demonstrate the abundant intracellular synthesis of single-atom zero-valent copper by this bacterium. This finding shows that microbes in acid mine drainages can naturally extract metal ions, such as copper, and transform them into a valuable commodity.

|
Scooped by
?
April 25, 2021 3:12 PM
|
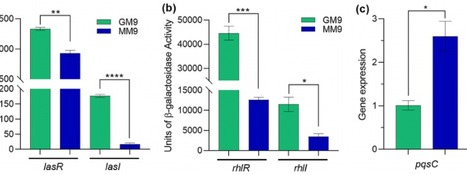
Pseudomonas aeruginosa is an opportunistic pathogen that uses malonate among its many carbon sources. We recently reported that, when grown in blood from trauma patients, P. aeruginosa expression of malonate utilization genes was upregulated. In this study, we explored the role of malonate utilization and its contribution to P. aeruginosa virulence. We grew P. aeruginosa strain PA14 in M9 minimal medium containing malonate (MM9) or glycerol (GM9) as a sole C source and assessed the effect of the growth on quorum sensing, virulence factors, and antibiotic resistance. Growth of PA14 in MM9, compared to GM9, reduced the production of elastases, rhamnolipids, and pyoverdine; enhanced the production of pyocyanin and catalase; and increased its sensitivity to norfloxacin. Growth in MM9 decreased extracellular levels of N‐acylhomoserine lactone autoinducers, an effect likely associated with increased pH of the culture medium; but had little effect on extracellular levels of PQS. At 18 h of growth in MM9, PA14 formed biofilm‐like structures or aggregates that were associated with biomineralization, which was related to increased pH of the culture medium. These results suggest that malonate significantly impacts P. aeruginosa pathogenesis by influencing the quorum sensing systems, the production of virulence factors, biofilm formation, and antibiotic resistance.
|
 Your new post is loading...
Your new post is loading...































increase natural DNA transformation in the strain O395.