 Your new post is loading...

|
Scooped by
?
Today, 6:46 PM
|
Identifying beneficial or functional mutations for a specific species is a task always relies on the labor-intensive construction of a library containing thousands of single-gene knockout or interference strains. Here, we systematically demonstrated that the task could be done by constructing a genome-scale library, designed by AutoGSL, containing a limited number of large fragment deletion strains. The loss-of- and gain-of-function phenotypes could be efficiently screened out by our chromosome-segment scanning, and the specific gene corresponding to an observed phenotype could be quickly identified and visualized by our computer-aided gene-annotation web tool. Additionally, a Clusters of Orthologous Gene Transformer learned representations were transferred to predict growth phenotypes of the genome-scale library under varying conditions. We further utilized our chromosome segment scanning for gain- or loss-of-function screening (CHASING) strategy to obtain acetoin- and lycopene-overproducing hosts. Our work highlighted the significance of CHASING in functional genomics investigation, robust chassis engineering, and chemical overproduction.

|
Scooped by
?
Today, 6:33 PM
|
Alternative food products are needed to address the most pressing challenges faced by the food industry: growing global food demand, health concerns, animal welfare, food security and environmental sustainability. Future foods are defined as foods with scalability and sustainability potential owing to rapidly advancing technological developments in their production systems. Key areas of study for future foods include cellular agriculture and plant-based systems, which include biomaterials as key ingredients or as structural components to impart texture, support cell growth and metabolism, and provide nutrients and organoleptic factors to food products. This Review discusses current requirements, options and processing approaches for biomaterials with utility in future foods. We focus on two main approaches: cellular agriculture wherein the cells are the key component for food (with the biomaterials utilized to support the cells via adherence and/or for texture) and plant-based foods wherein acellular plant-derived biomaterials are the food components. In both cases, the same fundamental challenges apply for the biomaterials: achieving utility at scale and low cost while meeting food safety requirements. Other considerations for biomaterials for future foods are also addressed, including sustainability, modelling, consumer acceptance, nutrition, regulatory status and safety considerations to highlight the path ahead. This emerging field of biomaterials for future foods offers a new generation of biomaterial systems that can positively impact human health, environmental sustainability and animal welfare. Although scaling these biomaterial sources cost-effectively presents a major challenge, substantial progress is being made, and opportunities to establish supply chains are already underway. Biomaterials have a crucial role in the development of future foods, particularly in cellular agriculture and plant-based systems. This Review addresses the current status and future requirements of biomaterials for future foods, addressing key aspects such as structure, nutrition, safety, sensory attributes, sustainability and consumer preferences.

|
Scooped by
?
Today, 2:16 AM
|
A key unresolved question in microbial ecology is how the extraordinary diversity of microbiomes emerges from the interactions among their many functionally distinct populations. This process is driven in part by the cross-feeding networks that help to structure these systems, in which consumers use resources to fuel their metabolism, creating by-products which can be used by others in the community. Understanding the effects of cross-feeding presents a major challenge, as it creates complex interdependencies between populations which can be hard to untangle. We address this problem using the tools of network science to develop a structural microbial community model. Using methods from percolation theory, we identify feasible community states for cross-feeding network structures in which the needs of consumers are met by metabolite production across the community. We identify tipping points at which small changes in structure can cause the catastrophic collapse of cross-feeding networks and abrupt declines in microbial community diversity. Our results are an example of a well-defined tipping point in a complex ecological system and provide insight into the fundamental processes shaping microbiomes and their robustness. We further demonstrate this by considering how network attacks affect community diversity and apply our results to show how the apparent difficulty in culturing the microbial diversity emerges as an inherent property of their cross-feeding networks.

|
Scooped by
?
Today, 2:05 AM
|
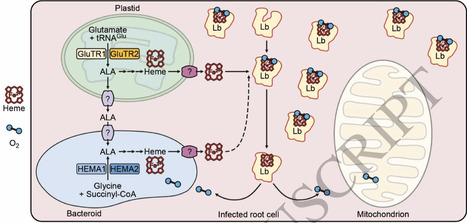
Heme is biosynthesized in legume root nodules to meet the demand for leghemoglobins (Lbs) and other heme-binding proteins. However, the main source of nodule heme remains unknown. Both the plant host and rhizobia possess a complete heme biosynthetic pathway, differing slightly in the production of 5-aminolevulinic acid (ALA), a key regulatory step catalyzed by glutamyl-tRNA reductase (GluTR) in the plant and by HemA in the rhizobia. Transcriptomic analysis revealed that many plant heme biosynthetic genes, including GluTR2 but not GluTR1, are upregulated in nodules compared to roots, whereas expression of related rhizobial genes, including both HemA1 and HemA2, is generally inhibited under symbiotic conditions compared to free-living conditions. Knockout of Lotus japonicus GluTR2, but not of HemA1 and HemA2, led to a significant decrease (∼50%) in nodule heme content. The stable heterozygous mutant of GluTR1 or transient knockdown of GluTR1 exhibited a ∼20% reduction in nodule heme content. Overexpression of Fluorescent in blue light (FLU), a feedback inhibitor of GluTR activity, caused a much greater reduction in nodule heme content (∼75%) and an increased level of apo-Lb and, in combination with the hemA1 hemA2 mutant, a drastic inhibition of nitrogenase activity (>90%). This study provides genetic evidence supporting a major role of plant GluTRs in coordinating heme biosynthesis between the two symbionts by supplying heme to assemble with cytoplasmic apo-Lbs and by providing ALA for heme synthesis in the bacteroids.

|
Scooped by
?
Today, 1:51 AM
|
Plant-associated microorganisms interact with each other and host plants via intricate chemical signals, offering multiple benefits, including plant nutrition. We report such a mechanism through which the rhizosphere microbiome improves plant growth under sulfur (S) deficiency. Disruption of plant S homeostasis resulted in a coordinated shift in composition and S-metabolism of the rhizosphere microbiome. Using this insight, we developed an 18-membered synthetic rhizosphere bacterial community (SynCom) that rescued growth of Arabidopsis and a leafy Brassicaceae vegetable under S-deficiency. This beneficial trait is taxonomically widespread among SynCom members, with bacterial pairs predominantly providing synergistic benefits to host plants. Notably, stronger competitive interactions among SynCom members conferred greater fitness benefits to the host, suggesting a trans-kingdom fitness tradeoff. Finally, guided chemical screening, deletion knockout mutant, and targeted metabolomics identified and validated microbially released glutathione (GSH) as the necessary bioactive signal that orchestrates plant-microbe (trans-kingdom) fitness tradeoff and improves plant growth under sulfur limitation.

|
Scooped by
?
Today, 1:37 AM
|
Phages are typically classified as temperate, integrating into host genomes, or lytic, replicating and killing bacteria. Lytic phages are not expected in bacterial genome sequences, yet our analysis of 3.6 million bacterial assemblies from 1,226 species revealed >100,000 complete lytic phage genomes, which we term Bacterial Assembly-associated Phage Sequences (BAPS). This represents a ~five-fold increase in the number of phages with known hosts and fundamentally reshapes our understanding of phage biology. Identifying BAPS has enabled the discovery of entirely novel phage clusters, including clusters distantly related to Salmonella Goslarviruses in E. coli, and Shigella, while significantly expanding known genera such as Seoulvirus (from 16 to >300 members). Notably, close relatives of therapeutic lytic phages were also detected, suggesting clinical isolate sequencing unknowingly archives viable phage candidates. The discovery of complete, lytic phage genomes within bacterial assemblies challenges long-standing assumptions and reveals a vast, untapped reservoir of phages.

|
Scooped by
?
Today, 1:31 AM
|
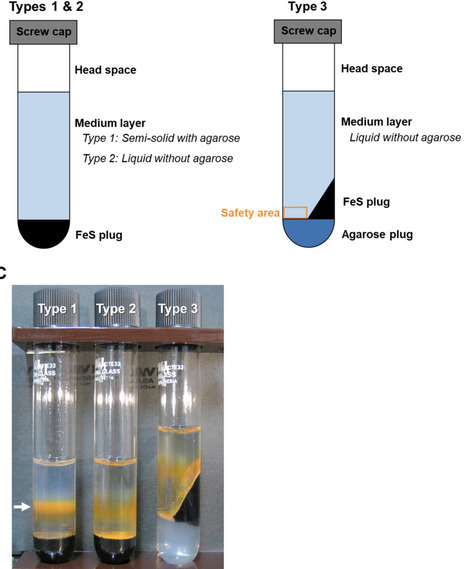
Chemolithotrophic neutrophilic iron (Fe)-oxidizing bacteria, which mainly belong to the family Gallionellaceae, universally prevail in terrestrial environments changing iron cycling. However, they are typically recognized as difficult-to-culture microbes. Despite efforts, there are few Fe(II)-oxidizing lithotroph isolates; hence, their physiological and ecological knowledge remains limited. This limitation is largely owing to difficulties in their cultivation, and we hypothesize that the difficulty exists because substrate and mineral concentrations in the cultivation medium are not tuned to a specific environmental condition under which these organisms live. To address this hypothesis, this study proposes a novel custom-made medium approach for chemolithotrophic Fe(II)-oxidizing bacteria; a method that manipulates medium components through diligent analysis of field environment. A new custom-made medium simulating energy substrates and nutrients under the field condition was prepared by modifying both chemical composition and physical setup in the glass-tube medium. In particular, the modification of the physical setup in the tube had a significant effect on adjusting dissolved Fe(II) and O2 concentrations to the field environment. Using the medium, Gallionellaceae members were successfully enriched and a new Gallionellaceae species was isolated from a natural hot spring site. Compared with conventional medium, the custom-made medium has significantly higher ability in enriching Gallionellaceae members.

|
Scooped by
?
Today, 1:23 AM
|
Metabolites are chemically heterogeneous and difficult to quantify in easily read formats. Recently, Tan and Fraser demonstrated that metabolites can be readily quantified by pairing aptamer function with DNA sequencing. This reflects a larger trend of sequencing for assessing biomolecule abundances, further leading to sequencing being a universal analytical tool.

|
Scooped by
?
Today, 1:20 AM
|
The development of sustainable and economically competitive biotechnological processes is a central challenge of modern industrial biotechnology. Conventional strategies such as macroscopic and molecular bioprocess design are often insufficient to exploit their full potential. To circumvent this, molecular process control provides the missing link to further consolidate various optimization strategies to achieve multilayered process design. This review highlights the molecular mechanisms that can be exploited for molecular process control. These can either be endogenous or specifically implemented into the organism, and comprise regulatory mechanisms at the transcriptional, translational, and system levels. In addition to serving as a design tool to enhance existing bioprocesses, molecular process control is the gateway to biotechnological advances that will extend the boundaries of future process design.

|
Scooped by
?
Today, 1:09 AM
|
Polyphenols are plant-derived secondary metabolites known for their antioxidants, anti-inflammatory, and antimicrobial properties, with flavonoids being the most structurally diverse and medically relevant subclass. Traditional plant extraction is limited by low abundance and difficulty in separating from analogs. Microbial synthesis has emerged as an alternative method to complement plant extraction. This review summarizes recent advancements in microbe-sourced polyphenols, especially flavonoids and related derivatives. Key strategies, including modular design, CRISPR-based optimization, co-culture, and dynamic regulatory systems, have been employed to enhance microbial factory production efficiency. Emerging artificial intelligence–driven computational modeling and pathway optimization hold significant promise for enhancing polyphenol biosynthesis. Taken together, microbial synthesis offers a scalable and sustainable alternative to plant extraction. The cost-effective production of polyphenols will expand their applications in pharmaceuticals, nutraceuticals, and food industry.

|
Scooped by
?
Today, 1:01 AM
|
Chemical gradients and the emergence of distinct microenvironments in biofilms are vital to the stratification, maturation and overall function of microbial communities. These gradients have been well characterized throughout the biofilm mass, but the microenvironment of recently discovered nutrient transporting channels in Escherichia coli biofilms remains unexplored. This study employs three different oxygen sensing approaches to provide a robust quantitative overview of the oxygen gradients and microenvironments throughout the biofilm transport channel networks formed by E. coli macrocolony biofilms. Oxygen nanosensing combined with confocal laser scanning microscopy established that the oxygen concentration changes along the length of biofilm transport channels. Electrochemical sensing provided precise quantification of the oxygen profile in the transport channels, showing similar anoxic profiles compared with the adjacent cells. Anoxic biosensing corroborated these approaches, providing an overview of the oxygen utilization throughout the biomass. The discovery that transport channels maintain oxygen gradients contradicts the previous literature that channels are completely open to the environment along the apical surface of the biofilm. We provide a potential mechanism for the sustenance of channel microenvironments via orthogonal visualizations of biofilm thin sections showing thin layers of actively growing cells. This complete overview of the oxygen environment in biofilm transport channels primes future studies aiming to exploit these emergent structures for new bioremediation approaches.

|
Scooped by
?
Today, 12:33 AM
|
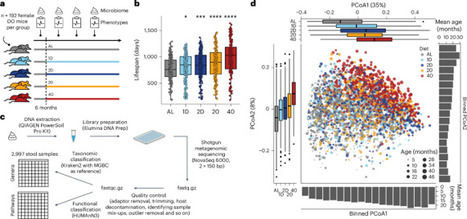
The gut microbiome changes with age and has been proposed to mediate the benefit of lifespan-extending interventions such as dietary restriction. However, the causes and consequences of microbiome aging and the potential of such interventions remain unclear. Here we analysed 2,997 metagenomes collected longitudinally from 913 deeply phenotyped, genetically diverse mice to investigate interactions between the microbiome, ageing, dietary restriction (caloric restriction and fasting), host genetics and a range of health parameters. Among the numerous age-associated microbiome changes that we find in this cohort, increased microbiome uniqueness is the most consistent parameter across a second longitudinal mouse experiment that we performed on inbred mice and a compendium of 4,101 human metagenomes. Furthermore, cohousing experiments show that age-associated microbiome changes may be caused by an accumulation of stochastic environmental exposures (neutral theory) rather than by the influence of an ageing host (selection theory). Unexpectedly, the majority of taxonomic and functional microbiome features show small but significant heritability, and the amount of variation explained by host genetics is similar to ageing and dietary restriction. We also find that more intense dietary interventions lead to larger microbiome changes and that dietary restriction does not rejuvenate the microbiome. Lastly, we find that the microbiome is associated with multiple health parameters, including body composition, immune components and frailty, but not lifespan. Overall, this study sheds light on the factors influencing microbiome ageing and aspects of host physiology modulated by the microbiome. Using a cohort of nearly 1,000 genetically diverse mice undergoing dietary restriction over the lifespan, the authors reveal numerous insights into the interactions between the gut microbiome, host genome, diet, health and longevity.

|
Scooped by
?
Today, 12:09 AM
|
The ability to detect and quantify microbiota over time from shotgun metagenomic data has a plethora of clinical, basic science and public health applications. Given these applications, and the observation that pathogens and other taxa of interest can reside at low relative abundance, there is a critical need for algorithms that accurately profile low-abundance microbial taxa with strain-level resolution. Here we present ChronoStrain: a sequence quality- and time-aware Bayesian model for profiling strains in longitudinal samples. ChronoStrain explicitly models the presence or absence of each strain and produces a probability distribution over abundance trajectories for each strain. Using synthetic and semi-synthetic data, we demonstrate how ChronoStrain outperforms existing methods in abundance estimation and presence/absence prediction. Applying ChronoStrain to two human microbiome datasets demonstrated its improved interpretability for profiling Escherichia coli strain blooms in longitudinal faecal samples from adult women with recurring urinary tract infections, and its improved accuracy for detecting Enterococcus faecalis strains in infant faecal samples. Compared with state-of-the-art methods, ChronoStrain’s ability to detect low-abundance taxa is particularly stark. ChronoStrain accurately profiles low-abundance strains in longitudinal samples by jointly modelling nucleotide sequencing errors, strain presence/absence and the temporal information associated with each sample using a differentiable Bayesian model.
|

|
Scooped by
?
Today, 6:37 PM
|
Studies of membrane protein folding have progressed from simple systems such as bacteriorhodopsin to complex structures such as ATP-binding cassette transporters and voltage-gated ion channels. Advances in techniques such as single-molecule force spectroscopy and in vivo force profiling now allow for the detailed examination of membrane protein folding pathways at amino acid resolutions. These proteins navigate rugged energy landscapes partly shaped by the absence of hydrophobic collapse and the viscous nature of the lipid bilayer, imposing biophysical limitations on folding speeds. Furthermore, many transmembrane (TM) helices display reduced hydrophobicity to support functional requirements, simultaneously increasing the energy barriers for membrane insertion, a manifestation of the evolutionary trade-off between functionality and foldability. These less hydrophobic TM helices typically insert and fold as helical hairpins, following the protein synthesis direction from the N terminus to the C terminus, with assistance from endoplasmic reticulum (ER) chaperones like the Sec61 translocon and the ER membrane protein complex. The folding pathways of multidomain membrane proteins are defined by allosteric networks that extend across various domains, where mutations and folding correctors affect seemingly distant domains. A common evolutionary strategy is likely to be domain specialization, where N-terminal domains enhance foldability and C-terminal domains enhance functionality. Thus, despite inherent biophysical constraints, evolution has finely tuned membrane protein sequences to optimize foldability, stability, and functionality.

|
Scooped by
?
Today, 6:25 PM
|
Nitrous oxide (N2O) is the third most important greenhouse gas and originates primarily from natural and engineered microbiomes. Effective emission mitigations are currently hindered by the largely unresolved ecophysiological controls of coexisting N2O-converting metabolisms in complex communities. To address this, we used biological wastewater treatment as a model ecosystem and combined long-term metagenome-resolved metaproteomics with ex situ kinetic and full-scale operational characterization over nearly 2 years. By leveraging the evidence independently obtained at multiple ecophysiological levels, from individual genetic potential to actual metabolism and emergent community phenotype, the cascade of environmental and operational triggers driving seasonal N2O emissions has ultimately been resolved. We identified nitrifier denitrification as the dominant N2O-producing pathway and dissolved O2 as the prime operational parameter, paving the way to the design and fostering of robust emission control strategies. This work exemplifies the untapped potential of multi-meta-omics in the mechanistic understanding and ecological engineering of microbiomes towards reducing anthropogenic impacts and advancing sustainable biotechnological developments. Nitrous oxide is one of the main greenhouse gases emitted during biological wastewater treatment. This long-term multi-meta-omics analysis of a wastewater treatment plant identifies the main causes of nitrous oxide emissions and actionable mitigation strategies.

|
Scooped by
?
Today, 2:08 AM
|
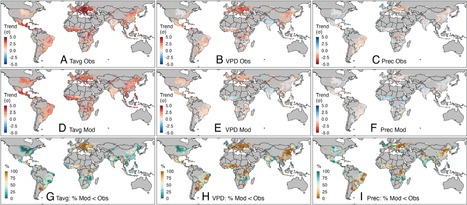
Efforts to anticipate and adapt to future climate can benefit from historical experiences. We examine agroclimatic conditions over the past 50 y for five major crops around the world. Most regions experienced rapid warming relative to interannual variability, with 45% of summer and 32% of winter crop area warming by more than two SD (σ). Vapor pressure deficit (VPD), a key driver of plant water stress, also increased in most temperate regions but not in the tropics. Precipitation trends, while important in some locations, were generally below 1σ. Historical climate model simulations show that observed changes in crops’ climate would have been well predicted by models run with historical forcings, with two main surprises: i) models substantially overestimate the amount of warming and drying experienced by summer crops in North America, and ii) models underestimate the increase in VPD in most temperate cropping regions. Linking agroclimatic data to crop productivity, we estimate that climate trends have caused current global yields of wheat, maize, and barley to be 10, 4, and 13% lower than they would have otherwise been. These losses likely exceeded the benefits of CO2 increases over the same period, whereas CO2 benefits likely exceeded climate-related losses for soybean and rice. Aggregate global yield losses are very similar to what models would have predicted, with the two biases above largely offsetting each other. Climate model biases in reproducing VPD trends may partially explain the ineffectiveness of some adaptations predicted by modeling studies, such as farmer shifts to longer maturing varieties.

|
Scooped by
?
Today, 1:56 AM
|
Polyhydroxyalkanoates are biopolyesters synthesized and stored in intracellular granules by diverse prokaryotes. Despite intense research efforts and prior evidence of a rather widespread phylogenetic occurrence of the related genetic machinery, reports on extreme thermophilic and hyperthermophilic polyhydroxyalkanoates producers remain scarce. However, thermophilic cell factories for bioplastic production would serve as an excellent example of Next-Generation Industrial Biotechnology. In this study, we aim to address this research gap by establishing a bioinformatics pipeline to mine genomes of extremely and moderately thermophilic microorganisms for signatures of potential polyhydroxyalkanoate PHB production. Based on a collection of verified protein sequences of polyhydroxyalkanoate polymerase PhaC, the key biosynthetic enzyme, carefully curated sets of thermophilic bacterial and archaeal genomes were screened. This revealed that although PhaC-encoding genes are prevalent in diverse moderately thermophilic bacteria, they are absent in the considered extreme thermophilic bacteria. In contrast, a few limited examples of extreme thermophilic archaea were found to encode putative phaC genes embedded within a typical polyhydroxyalkanoate synthesis operon in their genomes, namely within the genera Ferroglobus, Geoglobus and Archaeoglobus, while no hits were found in extreme thermophilic bacteria. The latter included Thermus thermophilus, which was previously reported as a polyhydroxyalkanoates producer. This was refuted in our bioinformatics analysis and moreover, the predicted absence of polyhydroxyalkanoates synthesis in T. thermophilus was experimentally confirmed by employing various extraction and analytical methods. Based on the findings in this study, we conclude that polyhydroxyalkanoate production is very scarce in extreme thermophiles and hyperthermophiles, for reasons that remain to be elucidated.

|
Scooped by
?
Today, 1:44 AM
|
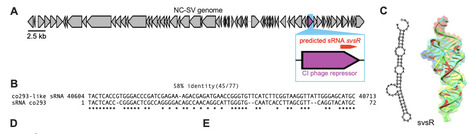
Prophages are prevalent features of bacterial genomes that can reduce susceptibility to lytic phage infection, yet the mechanisms involved are often elusive. Here, we identify a small RNA (svsR) encoded by the lambdoid prophage NC-SV in adherent-invasive Escherichia coli (AIEC) strain NC101 that confers resistance to lytic coliphages. Comparative genomic analyses revealed that NC-SV-like prophages and svsR homologs are conserved across diverse Enterobacteriaceae. Transcriptional analyses reveal that svsR represses maltodextrin transport genes, including lamB, which encodes the outer membrane maltoporin LamB--a known receptor for multiple phages. Nutrient supplementation experiments show that maltodextrin enhances phage adsorption, while glucose suppresses it, consistent with established effects of these sugars on lamB expression. In vivo, we compared wild-type NC101 and a prophage-deletion strain (NC101deltaNC-SV) in mice to assess the impact of NC-SV on lytic phage susceptibility. Although intestinal E. coli densities remained stable across groups, animals colonized with NC101 exhibited markedly reduced phage burdens in both the intestinal lumen and mucosa compared to mice colonized with NC101deltaNC-SV. This reduced phage pressure was associated with increased dissemination of NC101 to extraintestinal tissues, including the spleen and liver. Together, these findings highlight a nutrient-responsive, prophage-encoded mechanism that protects AIEC from phage predation and may promote bacterial persistence and dissemination in the inflamed gut.

|
Scooped by
?
Today, 1:35 AM
|
Over the past two decades, metagenomics has greatly expanded our understanding of microbial phylogenetic and metabolic diversity. However, most microbial taxa remain uncultured, hindering research and biotechnological applications. Isolating environmental anaerobes using traditional methods is particularly cumbersome and low throughput. Here, we present a novel, high-throughput approach for the cultivation and isolation of anaerobes, which involves trapping and growing single microbes within selectively permeable hydrogel capsules followed by fluorescence-activated cell sorting to distribute compartmentalized isolates into liquid medium for further growth. We show that diverse anaerobes can grow within capsules and that slower-growing ones (e.g. methanogens) can be enriched with this platform. We also applied this approach to isolate anaerobes from soil, including strains of the sulfate-reducing bacteria Desulfovibrio desulfuricans and Nitratidesulfovibrio vulgaris. Overall, this work introduces a robust, high-throughput alternative to traditional techniques for isolating environmental anaerobes and expands the emerging set of microfluidics-based tools for the cultivation of novel taxa.

|
Scooped by
?
Today, 1:27 AM
|
Advances in precise genome editing are enabling genomic recordings of cellular events. Since the initial demonstration of CRISPR-based genome editing, the field of genomic recording has witnessed key strides in lineage recording, where clonal lineage relationships among cells are indirectly recorded as synthetic mutations. However, methods for directly recording and reconstructing past cellular events are still limited, and their potential for revealing new insights into cell fate decisions has yet to be realized. The field needs new sensing modules and genetic circuit architectures that faithfully encode past cellular states into genomic DNA recordings to achieve such goals. Here we review recently developed strategies to construct diverse sensors and explore how emerging synthetic biology tools may help to build molecular circuits for genomic recording of diverse cellular events.

|
Scooped by
?
Today, 1:20 AM
|

|
Scooped by
?
Today, 1:12 AM
|
Optogenetics has emerged as a powerful tool for regulating cellular processes due to its noninvasive nature and precise spatiotemporal control. Far-red light (FRL) has increasingly been used in the optogenetic control of mammalian cells due to its low toxicity and high tissue penetration. However, robust and orthogonal FRL sensors are lacking in bacteria. Here, we established an orthogonal FRL sensor in Escherichia coli with a maximum dynamic range exceeding 230-fold based on the RfpA-RfpC-RfpB (RfpABC) signaling system that regulates the far-red light photoacclimation (FaRLiP) in cyanobacteria. We identified a conserved DNA motif in the promoter sequences of the Chl f synthase gene and other genes in the FaRLiP gene clusters, termed the far-red light-regulatory (FLR) motif, which enables the light-responsive activation of gene expression through its interaction with RfpB. Based on the FLR motif, we simplified the FLR-containing promoters and characterized their activation abilities and dynamic ranges, which can be utilized in different synthetic biology scenarios. Additionally, one or two FLR motifs are present at other loci within the FaRLiP gene cluster, providing further FRL-inducible promoter resources. The FRL sensor exhibits effective activation and suppression under low-intensity FRL and white light, respectively, and remains functional in darkness. In conclusion, this study advances the understanding of the regulatory mechanisms of FaRLiP in cyanobacteria and provides robust and orthogonal FRL sensors for synthetic biology applications.

|
Scooped by
?
Today, 1:06 AM
|
One-carbon (C1) substrates are attractive feedstocks for biological upgrading as part of a circular, carbon-negative bioeconomy. Nature has evolved a diverse set of C1-trophs that use a variety of pathways. Additionally, intensive effort has recently been invested in developing synthetic C1 assimilation pathways. This complicated landscape presents the question: “What pathways should be used to produce what products from what C1 substrates?” To guide the selection, we calculate and compare maximal theoretical yields for a range of bioproducts from different C1 feedstocks and pathways. The results highlight emerging opportunities to apply metabolic engineering to specific C1 pathways to improve pathway performance. Since the C1 landscape is dynamic, with new discoveries in the biochemistry of native pathways and new synthetic alternatives rapidly emerging, we present detailed procedures for these yield calculations to enable others to easily adapt them to additional scenarios as a foundation for establishing industrially relevant production strains.

|
Scooped by
?
Today, 12:55 AM
|
The bacterial genus Massilia was first described in 1998, and since then has attracted growing interest due to its ecological plasticity and biotechnological promise. Certain species of the genus Massilia inhabit a variety of ecosystems, from arid deserts to polar glaciers, and exhibit unique adaptations such as resistance to cold and heat. In contaminated environments, some members of Massilia contribute significantly to the detoxification of heavy metals and the degradation of organic pollutants, presenting them as promising agents for bioremediation. In addition, Massilia species improve plant resistance and facilitate pollutant absorption in phytoremediation strategies. New research also highlights their potential as bioindicators of environmental health, given their abundance in anthropogenically influenced ecosystems and airborne microbial communities. In addition to their ecological roles, some Massilia species have potential in biotechnological applications by producing biopolymers and secondary metabolites. Here, we integrate findings across various habitats to present a comprehensive overview of the ecological and biotechnological importance of the genus Massilia. We highlight critical knowledge gaps and propose future research directions to fully harness the potential of this not fully explored bacterial genus to address environmental challenges, including contamination.

|
Scooped by
?
Today, 12:16 AM
|
Bacteriophages show promise for microbiome engineering, but studying their transmission dynamics in multimember communities and animal hosts is technically challenging. We therefore created ‘Phollow’, a live imaging-based approach for tracking phage replication and spread in situ with single-virion resolution. Following interbacterial phage transmission is achieved by marking virions with distinct fluorescent proteins during assembly in newly infected cells. In vitro cell virology studies revealed clouds of phage virions dispersing upon bacterial lysis, leading to rampant transmission. Combining Phollow with optically transparent zebrafish, we visualized phage outbreaks within the vertebrate gut. We observed that virions from a zebrafish-derived Plesiomonas strain, but not a human-derived E. coli, rapidly disseminate systemically to the liver and brain. Moreover, antibiotics triggered waves of interbacterial transmission and sudden shifts in gut community ecology. Phollow ultimately empowers multiscale investigations of phage transmission and transkingdom interactions that have the potential to open new avenues for phage-based microbiome therapies. ‘Phollow’ is a live imaging-based fluorescence tagging approach that can track phage replication and spread in situ with single-virion resolution.
|
 Your new post is loading...
Your new post is loading...





























omv