 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
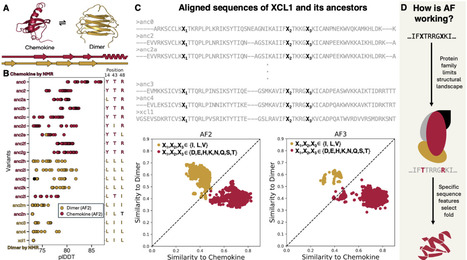
Transformer models, neural networks that learn context by identifying relationships in sequential data, underpin many recent advances in artificial intelligence. Nevertheless, their inner workings are difficult to explain. Here, we find that a transformer model within the AlphaFold architecture uses simple, sparse patterns of amino acids to select protein conformations. To identify these patterns, we developed a straightforward algorithm called Conformational Attention Analysis Tool (CAAT). CAAT identifies amino acid positions that affect AlphaFold's predictions substantially when modified. These effects are corroborated by experiments in several cases. By contrast, modifying amino acids ignored by CAAT affects AlphaFold predictions less, regardless of experimental ground truth. Our results demonstrate that CAAT successfully identifies the positions of some amino acids important for protein structure, narrowing the search space required to make effective mutations and suggesting a framework that can be applied to other transformer-based neural networks.

|
Scooped by
mhryu@live.com
Today, 12:45 AM
|
The microbiome is increasingly recognized as a key player in cancer pathogenesis and treatment response, acting through both local and systemic mechanisms. Microbial communities and their metabolites can directly influence drug metabolism, shape the immune landscape, and alter transcriptional and epigenetic programmes in the gut, systemically and in the tumor microenvironment. Emerging data support the potential of microbiome-targeted interventions (such as faecal microbiota transplantation, diet, prebiotics and probiotics) as adjuncts to conventional cancer therapies, with the goal of enhancing efficacy and reducing toxicity. This Review highlights the promise of the microbiome as a prognostic and predictive biomarker, a modifiable factor in cancer care and prevention, and a therapeutic target. We also discuss major knowledge gaps, limitations in current study designs, and the need for mechanism-guided, personalized strategies to advance clinical translation. In this Review, Hajjar, Mars and Kashyap discuss the role of the microbiome in cancer development and progression and in treatment response. They also consider the potential of the microbiome as a therapeutic target and as a prognostic and predictive biomarker, and outline major knowledge gaps in the field.

|
Scooped by
mhryu@live.com
January 4, 11:12 AM
|
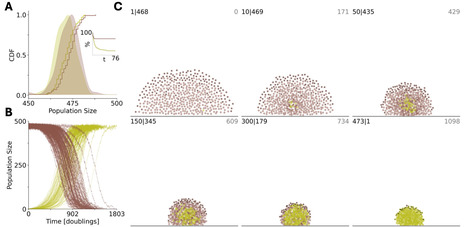
Evolution of cooperation is a major, extensively studied problem in evolutionary biology. Cooperation is beneficial for a population as a whole but costly for the bearers of social traits such that cheaters enjoy a selective advantage over cooperators. Here, we focus on coevolution of cooperators and cheaters in a multi-level selection framework, by modeling competition among groups composed of cooperators and cheaters. Cheaters enjoy a reproductive advantage over cooperators at the individual level, independent of the presence of cooperators in the group. Cooperators carry a social trait that provides a fitness advantage to the respective groups. In the case of absolute fitness advantage, where the survival probability of a group is independent of the composition of other groups, the survival of cooperators does not correlate with the presence of cheaters. By contrast, in the case of relative fitness advantage, where the survival probability of a group depends on the composition of all groups, the survival of cooperators positively correlates with the presence of cheaters. Increasing the strength of the social trait alone fails to ensure survival of cooperators, and the increase of the reproduction advantage of the cheaters is necessary to avoid population extinction. This unexpected effect comes from multilevel selection whereby cheaters at the individual level become altruists at the group level, enabling overall growth of the population that is essential for the persistence of cooperators. We validate these theoretical results with an agent-based model of a bacterial biofilm where emergence of the cooperative trait is facilitated by the presence of cheaters, leading to evolution of new spatial organization. Our results suggest that, counterintuitively, cheaters often promote, not destabilize, evolution of cooperation.

|
Scooped by
mhryu@live.com
January 4, 10:42 AM
|
The deep terrestrial subsurface (DTS) biosphere consists of a variety of distinct microbial taxa, mostly bacterial. The mechanisms by which microbes dynamically manage the uptake and concurrent utilization of nutrients within the DTS environments remain largely unexplored. Here, we examined the utilization patterns of amino acids and other polar metabolites in cultured DTS bacterial communities to investigate the adaptive responses and metabolic pathways employed under varying nutrient conditions to gain insight into how environmental shifts impact the metabolism of these communities. Previously, we found that changes in growth conditions affected the composition and size of the bacterial communities enriched from these oligotrophic, anoxic environments and induced changes in the production of primary and secondary metabolites. In present study, metabolic fingerprinting was used to investigate the primary and secondary metabolite utilization and main metabolic pathways present in the enriched DTS bacterial consortium originating from the deep Fennoscandian Shield. We found that especially amino acids were predominantly degraded under different nutrient conditions. Notably, the degradation of phenylalanine and valine constituted a 'core' metabolic process that remained unaffected by variations in available nutrients within this community. Further, the most significant metabolic pathways employed were those connected to phenylalanine, cysteine and methionine.

|
Scooped by
mhryu@live.com
January 4, 1:05 AM
|
Biological systems are inherently complex and heterogeneous. Deciphering this complexity increasingly relies on high-throughput single-cell omics methods and tools that efficiently probe the cellular phenotype and genotype. Here we present a versatile technology based on semipermeable capsules (SPCs), tailored for a variety of high-throughput nucleic acid assays, including single-cell genome and mRNA sequencing, and fluorescence-activated cell sorting-based isolation of individual transcriptomes based on nucleic acid marker of interest. Being biocompatible, the SPCs support single-cell cultivation and clonal expansion over long periods of time thereby overcoming a fundamental limitation of droplet microfluidics platforms. Overall, the SPCs represent customizable and broadly applicable tool for easy-to-use, scalable single-cell omics applications that are built on multi-step biochemical reactions.

|
Scooped by
mhryu@live.com
January 4, 12:50 AM
|
A more comprehensive understanding of the bacterial species in the vaginal microbiome and their roles requires their cultivation as pure isolates. However, difficulty in culturing fastidious species, particularly in liquid media, has limited certain types of experimental approaches for studying these organisms. To address this challenge, Vaginal Microbe Medium (VMM) was developed to enable robust growth of Lactobacillus iners within a relatively short period (36 h). A simplified version with fewer components, named VMM2, was subsequently developed. L. iners strains grown in VMM and VMM2 reached higher optical densities in a shorter period compared to NYCIII, supplemented MRS and Columbia broth. Bacterial cells cultured in VMM and VMM2 were significantly longer than those grown in NYCIII. These results indicate that VMM and VMM2 provide superior growth conditions for L. iners as compared to currently available media. Both media also support the growth of a range of other vaginal bacterial species. The development of these media facilitates liquid culture of fastidious bacteria from the vaginal niche, enabling broader experimental capabilities and deeper insights into causal relationships within the microbiome.

|
Scooped by
mhryu@live.com
January 4, 12:39 AM
|
E. coli employs diverse strategies to adapt to acidic environments that disrupt enzyme activity and the thermodynamic feasibility of essential reactions. To understand the impact of pH stress on cell metabolism, we present the PET-FBA (pH-, Enzyme protein allocation-, and Thermodynamics-constrained Flux Balance Analysis) framework. PET-FBA extends genome-scale modeling by integrating enzyme protein costs and reaction Gibbs free energy changes. Additionally, by incorporating pH-dependent enzyme kinetics in response to intracellular acidification, this framework enables the simulation of E. coli's metabolic adjustments across varying external pH levels. The model's accuracy is validated by comparing in silico growth simulations with experimental measurements under both anaerobic and aerobic conditions, as well as in silico gene knockouts of essential genes. By explicitly incorporating pH effects, our model accurately replicates the metabolic shift towards lactate production as the primary fermentation product at low pH in anaerobic conditions. This shift is only predicted when enzyme kinetics are dynamically adjusted as a function of pH. Further analysis revealed that this shift can be attributed to the reduced protein efficiency of the acetyl-CoA branch compared to lactate dehydrogenase under acidic stress, which then becomes crucial for maintaining NAD regeneration and cell growth at low pH. Furthermore, we i dentified strategies for enhancing cell growth under acidic anaerobic conditions by improving the enzyme activity of lactate dehydrogenase and pyruvate formate lyase, which increases NAD production efficiency and reduces enzyme protein allocation costs. Designed as a lightweight yet versatile framework, PET-FBA enables efficient genome-scale metabolic analysis. Using E. coli as a model system, our framework provides a systematic approach to understanding metabolic responses to environmental stress, pinpointing key metabolic bottlenecks, and identifying potential targets for strain optimization. This work also highlights the critical role of intracellular acidification in shaping enzyme performance and microbial adaptation. The PET-FBA framework is implemented as a Python package at https://github.com/Chaowu88/etfba, with detailed documentation provided at https://etfba.readthedocs.io.

|
Scooped by
mhryu@live.com
January 4, 12:26 AM
|
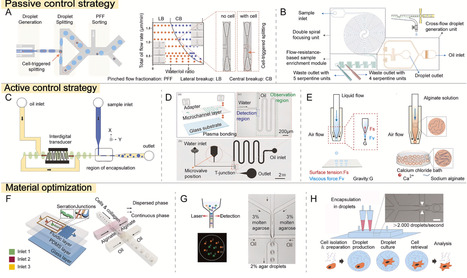
Single-cell analysis affords a novel perspective for deciphering the intricacies of complex biological systems by elucidating the subtle heterogeneities within cellular populations, with particular relevance to cellular development, pathophysiological mechanisms, and therapeutic responsiveness. At present, numerous studies have reported the application of droplet microfluidics in facilitating high-throughput experimentation at the single-cell level through the precise manipulation of individual cells. However, few comprehensive and systematic reviews have focused on the optimization of droplet microfluidic manipulation strategies and the innovative applications of this technology across various fields. This review discusses the ingenious designs for enhancing cell analysis and highlights their applications in the realms of bioassay, immunotherapy, and drug screening. Furthermore, this review summarizes the current research findings on droplet microfluidics and outlines their future development directions.

|
Scooped by
mhryu@live.com
January 4, 12:12 AM
|
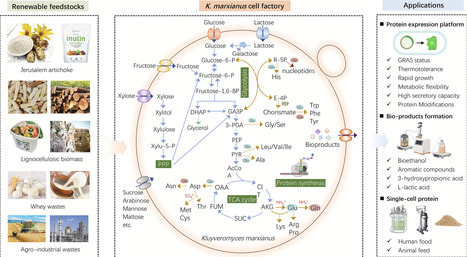
The non-conventional yeast Kluyveromyces marxianus is emerging as a versatile food-grade host with exceptional potential in industrial biotechnology, due to its rapid growth, thermotolerance, and metabolic flexibility. Its broad substrate utilization capacity and GRAS status have spurred growing interest in its application for recombinant protein expression and single-cell protein (SCP) production. However, a comprehensive understanding of the functional genomics and metabolic networks in K. marxianus remains limited. Nevertheless, ongoing efforts to develop diverse genetic engineering tools for K. marxianus have greatly strengthened its potential as a promising platform. In this review, we provide an extensive overview of the genetic, physiological, and biotechnological characteristics that establishes K. marxianus as an ideal host for efficient protein expression and SCP production. Recent advances and representative examples of engineering strategies aimed at unlocking its industrial potential for bioproduction were discussed. Finally, this review highlights the challenges and future directions for biotechnological innovations.

|
Scooped by
mhryu@live.com
January 3, 11:52 PM
|
Natural products (NPs) are increasingly applied in food, medicine, and biotechnology. However, their biosynthesis remains constrained by low titers and yields. Transcription factor-based biosensors (TFBs) can convert biological signals into measurable outputs, enabling real-time monitoring and dynamic regulation of biosynthetic pathways, thereby facilitating the overcoming of these limitations. This review highlights recent advances in applying TFBs to NP production, with a focus on high-throughput screening, adaptive evolution, and dynamic control. We further discuss innovative engineering approaches aimed at optimizing TFB performance, including in silico TF identification, protein engineering, and fine-tuning of regulatory elements. Finally, we examine the challenges associated with using TFBs for microbial NP production and explore their potential in emerging platforms such as cell-free systems and non-model microorganisms. These insights offer valuable perspectives on overcoming the current limitations of biosensing technologies and advancing the scalable production of NPs.

|
Scooped by
mhryu@live.com
January 3, 11:38 PM
|
The projected expansion of the global market for cell manufacturing, which contributes to regenerative medicine and cell therapies, warrants the designing and development of scalable cryopreservation processes for cell-based products (CBPs) for use in both standard and personalized therapies. However, the change in scale causes variations in process parameters, which affects the stability of the CBP quality. Therefore, the cryopreservation process for CBPs needs to be designed based on the concept of cell manufacturability and consideration of both engineering and biological aspects. In this review, we discussed strategies to enhance the quality stability of CBPs during cryopreservation, focusing primarily on four key processes: dispensing, freezing, storage, and thawing. Additionally, we discussed the application of simulation technologies because they aid in constructing digital twins for the designing and development of the cryopreservation process and facilitate efficiency with limited time and resources.

|
Scooped by
mhryu@live.com
January 3, 4:29 PM
|
Genome-scale metabolic network (GSMN) models are rigorously curated, cellular-level representations of metabolism that enable flux-based metabolite fate discovery, metabolic engineering, drug target identification and context-specific multi-omics integration. However, the inherent complexity of model architectures, need for programming skills and limited visualization support restrict their broader applicability. Existing tools focus on visualization and analyses separately, necessitating tool-specific format conversions, offer either topology or flux analyses options exclusively, lack intuitive pathway-specific visualizations, database-integrated model refinement, pathway enrichment and large-scale perturbation analyses. Here, we present NAViFluX (metabolic Network Analysis and Visualization of Flux), a visualization-centric, web browser-based environment that unifies GSMN exploration through native pathway/subsystem map generation using various layouts; interactive refinement through addition of reactions from KEGG/BiGG, amending constraints, pathway merging; and modules for flux computations, topology, functional enrichment. Instead of treating visualization as a post hoc step, NAViFluX performs all actions within network views, enabling users to make decisions directly from visually interpretable subnetworks. Using three Escherichia coli case studies, NAViFluX is shown to characterize nutrient-specific metabolic adaptations, improve metabolic gene essentiality predictions, provide mechanistic insight into synthetic lethality and facilitate the design of an optimized carbon-fixing metabolic state, thereby democratizing GSMNs for biologically meaningful applications.

|
Scooped by
mhryu@live.com
January 3, 4:14 PM
|
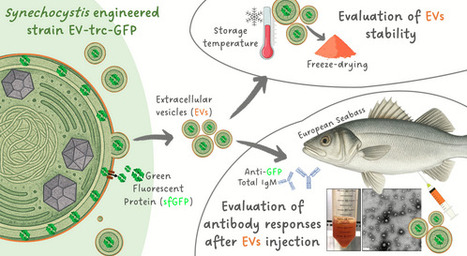
Fish aquaculture faces significant economic losses from disease outbreaks. Vaccination is the most effective prevention strategy, and bacterial extracellular vesicles (EVs) show promise as vaccine platforms due to their strong immuno-stimulating properties. However, the application of EVs derived from pathogenic bacteria is limited by toxicity risks and production challenges. Alternatively, genetic engineering of non-pathogenic microorganisms is being explored to produce tailored EVs to deliver antigens and serve as carriers of therapeutic proteins. Recently, we have engineered the model cyanobacterium Synechocystis sp. PCC 6803 for the expression of the reporter green fluorescent protein (sfGFP) and its targeting to EVs. Here, taking advantage of the Synechocystis sfGFP-loaded EVs, the stability of vesicles and their cargo was evaluated in the long term when stored under different temperature conditions and after freeze-drying. The possibility of using Synechocystis EVs as a tool for eliciting specific/adaptive immune responses was assessed in European seabass, a high commercial value fish, by following the amount of total and sfGFP-specific immunoglobulins produced after immunisation through injection. Synechocystis EVs were shown to be resilient nanostructures that can induce specific immune responses in fish with additional adjuvant features. This represents a biotechnological breakthrough towards a novel antigen-carrier platform for sustainable fish-pathogen control.
|

|
Scooped by
mhryu@live.com
Today, 12:57 AM
|
Molecular docking is a powerful computational tool for predicting protein-ligand interactions, widely employed in drug discovery. However, its effectiveness is often constrained by the availability of experimentally resolved X-ray protein structures, a process that is both time consuming and resource-intensive. AlphaFold (AF), a deep learning method, offers an efficient alternative by predicting high-accuracy 3D protein structures directly from amino acid sequences. This study assesses the utility of AF-generated protein models for fragment and larger ligand docking with Glide, a widely used docking approach. The docking workflow is evaluated in an unbiased manner by carrying out binding site identification with FTMap, a binding hot spot prediction software. We show that fragment docking to AF models outperforms docking to the respective unbound protein crystal structures, and performs comparably to docking to the corresponding ligand bound structures when using an unbiased approach. Leveraging computational efficiency of AF model generation, we also employ ensembles of AF models to incorporate protein flexibility. Results show that docking to AF ensembles improves larger-ligand docking compared to docking to singular AF models and outperforms docking to unbound structures. The results provide insights into the effectiveness of integrating AF protein models into docking procedures, highlighting the potential for streamlining computational drug discovery processes.

|
Scooped by
mhryu@live.com
January 4, 11:46 PM
|
Designing regulatable promoters with specified functional output remains difficult because natural promoters are unlikely to match a particular specification, and the sequence design space is large, complex, and challenging to interpret. This review advances a context-minimized, measurement-first approach in Escherichia coli that couples simple assays to a single transcription factor (TF)-based thermodynamic framework. The model is structured around two key concepts related to the TF: occupancy and function. Here, we outline how these concepts can be manipulated and measured at the level of DNA sequence and how those perturbations can impact fold-change and thus features of the promoter, such as dynamic range, leakiness, and sensitivity. LacI serves as a worked example in which sequence–occupancy, copy number, and competition, position-dependent function, and inducer allostery have been measured and can be combined to optimize response features. Overall, simple measurements linked to interpretable models provide a practical route to compiling desired regulatory specifications into sequence-level designs.

|
Scooped by
mhryu@live.com
January 4, 11:02 AM
|
The release of genetically engineered microorganisms into the environment has remained one of the most controversial and least explored frontiers in biotechnology. More than four decades after the early debates on the risks of recombinant DNA technology, field deployment of engineered bacteria to eliminate toxic waste and industrial and urban emissions remains practically frozen, particularly in Europe. It is one thing to engineer a strain with an environmentally useful trait—whether enhanced CO₂ fixation, xenobiotic breakdown, or explosives detection—and grow it in a Petri dish or a small bioreactor, and quite another to deliver that function effectively and safely at a scale required to address environmental problems.

|
Scooped by
mhryu@live.com
January 4, 1:15 AM
|
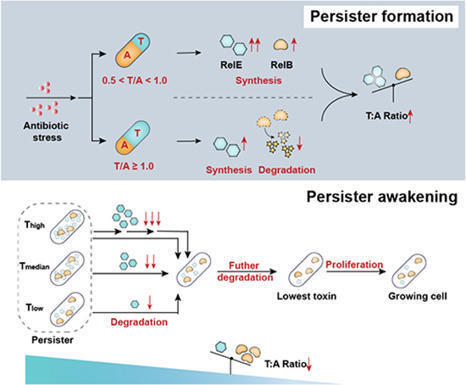
Persisters represent a transient, antibiotic-tolerant subpopulation within isogenic bacterial populations, contributing to infection relapses. However, the mechanisms driving persister formation and resuscitation remain elusive. Here, we developed nano-flow cytometry (nFCM)-based methods for single-cell quantification of toxin (T) RelE and antitoxin (A) RelB levels, as well as for monitoring persister states through cell wall growth. We demonstrate that bacteria elevate the T/A ratio through two distinct TA expression modalities to withstand bacteriostatic antibiotic challenge, with T/A = 1.0 as a critical threshold. Intriguingly, single-cell resuscitation dynamics revealed that subinhibitory antibiotic exposure promotes entry into a deeper dormant state characterized by elevated T/A ratios, underscoring the importance of maximizing therapeutic antibiotic concentrations. Crucially, we uncovered a triphasic detoxification process during resuscitation where progressive toxin depletion drives T/A ratio reduction to a critical proliferation-permissive threshold. Proteomic profiling unveiled that persisters with high RelE toxin production have increased transmembrane transporter levels linked to stress response and drug efflux. Our findings offer pivotal molecular insights underlying persister transitions and underscore the need for high-throughput, single-cell analysis of these heterogeneity phenotypes.

|
Scooped by
mhryu@live.com
January 4, 1:00 AM
|
Androgen receptor (AR) signaling is essential for prostate cancer (PCa) cell growth and remains a key therapeutic target in castration-resistant PCa (CRPC). While circular RNAs (circRNAs) are increasingly recognized as important regulatory molecules, their roles in AR signaling during PCa progression remain poorly understood. This study identified circUTRN, an AR-inhibited circRNA that is upregulated following neoadjuvant hormonal therapy and downregulated in PCa tissues. circUTRN inhibits proliferation in both castration-sensitive and castration-resistant PCa. Mechanistically, circUTRN binds to acetyl-CoA carboxylase 1 (ACC1) and impairs the activity through both phosphorylation-dependent and independent pathways, thereby disturbing de novo fatty acid synthesis. The dynamic relation between circUTRN and ACC1 expression during PCa progression from treatment-naïve to therapeutic-resistant states highlights the metabolic vulnerability of fatty acid synthesis. Notably, we developed nanoparticles to deliver circUTRN in combination with AR signaling inhibitors (ARSIs). This approach effectively suppressed CRPC xenograft tumor growth, even in models resistant to next-generation ARSIs. This study reveals an AR-regulated circRNA involved in PCa progression and suggests a potential therapeutic strategy for treatment-resistant PCa.

|
Scooped by
mhryu@live.com
January 4, 12:46 AM
|
Gene expression regulatory elements (GEREs) play a pivotal role in the control of gene transcription and translation. The design of GEREs with precise and tunable activity remains a major challenge in synthetic biology. Over the past decades, engineering strategies have evolved from empirical sequence mining and random mutagenesis to increasingly rational approaches guided by biophysical models and artificial intelligence. In this review, we systematically examine the design principles, representative studies, and implementation strategies for each GERE class, highlighting how mining, modular recombination, targeted mutagenesis, and deep generative modeling contribute to the development of functional regulatory elements. We further discuss the strengths and limitations of these strategies, offering practical guidance for optimizing microbial cell factory bioproduction through the fine-tuning of gene expression.

|
Scooped by
mhryu@live.com
January 4, 12:32 AM
|
Balancing metabolic pathways is critical for engineering microbial platforms to efficiently and robustly synthesize value-added bioproducts. In the oleaginous yeast Yarrowia lipolytica engineered for β-carotene production, lipid synthesis supports carotenoid storage but also competes with carotenoid synthesis for cellular resources, necessitating precise regulation for optimal resource allocation. In this study, we establish a machine learning framework that captures the complex interactions among three key metabolic modules for β-carotene synthesis: the mevalonate pathway (precursor supply for β-carotene), lipid synthesis (storage capacity), and the β-carotene synthetic cluster (product formation). This computational framework enables the prediction of β-carotene output based on gene combinations and guides iterative gene integration strategies across these interconnected pathways to optimize production. Using this approach, the best-performing strain YLT226 achieved a 7-fold increase in β-carotene titer compared to the initial strain YLT001 through nine rounds of guided gene integration. This work provides a promising strategy for understanding and engineering metabolic flux distributions.

|
Scooped by
mhryu@live.com
January 4, 12:18 AM
|
The field of synthetic biology is essential to the continued development of a bio-based economy, creating mechanisms to supply carbon needed in the economy by both converting existing end-of-life wastes as well as by creating novel, purpose-grown and sustainable feedstocks. Here, we first discuss the near- and long-term resources available for use as feedstocks for bioconversion as well as the output molecules needed for building the foundation of an expanded bio-based economy. We then outline the organisms and phenotypic traits that are needed for the performance-advantaged chassis organisms of the future. Furthermore, we detail the advances, challenges, and opportunities in both microbial and plant synthetic biology relevant to expanding the bio-based economy. Finally, we explore technologies that have and will further enable advances in synthetic biology and the greater bio-based economy.

|
Scooped by
mhryu@live.com
January 4, 12:10 AM
|
Lignin biosynthesis and plant cell wall engineering are central to plant structural integrity and biomass utility. Recent advances in molecular and synthetic biology have opened opportunities to tailor lignin contents, composition, and polymer structure for renewable bioenergy and sustainable biomaterial applications. This review provides an integrative perspective on biosynthesis, regulation, and engineering of lignin. It summarizes the current progress in understanding the genetic, transcriptional, epigenetic, and metabolic networks that control lignin formation, with a focus on emerging tools such as CRISPR/Cas genome editing, synthetic promoters, and metabolic rewiring. Beyond cataloguing current knowledge, it critically analyzes the trade-offs involved in lignin modification for biomaterials, addressing unresolved challenges such as monolignol transport, metabolic flux control, and species-specific regulatory divergence. Engineered lignin and modified plant cell walls hold significant potential for biorefineries, advanced polymers, pharmaceuticals, and carbon sequestration, yet their translation from the laboratory to the field remains limited. Engineered lignin offers real-world applications across diverse industries, including bioenergy, bioplastics, carbon fiber composites, pharmaceuticals, and sustainable construction materials, thereby reinforcing its pivotal role in advancing a circular bioeconomy. The review further proposes future research directions that integrate multi-omics, single-cell technologies, machine learning, and field-based validation to enable precision lignin engineering. Strategic advances in this field will support next-generation bioenergy systems, advanced biomaterials, and the transition to a circular bioeconomy.

|
Scooped by
mhryu@live.com
January 3, 11:45 PM
|
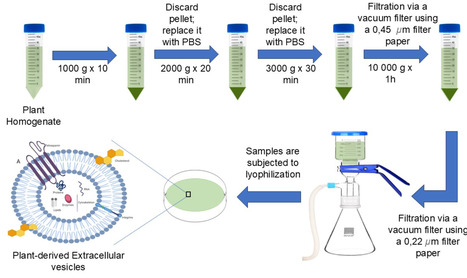
A major challenge in central nervous system disorders such glioblastoma includes the presence of a blood-brain barrier which restricts the delivery of therapeutic agents to the brain, thereby limiting the effectiveness of most conventional treatments. Moreover, the discovery of novel drugs for glioblastoma has been limited hence drug repurposing has gained traction leveraging existing drugs like itraconazole. Plant-derived extracellular vesicles (PDEVs) have potential as a natural pharmaceutical delivery system owing to their therapeutic capabilities. These PDEVs may be a good candidate for blood-brain barrier permeation due to their biomolecular composition and high drug loading efficiency of itraconazole. In this work, PDEVs isolated from aloe aborescens (aloe), Zingiber officinale (ginger) and Nigella sativa seeds [black cumin seeds (BCS)] were compared in terms of their physicochemical properties, drug release kinetics, cytotoxicity, cellular uptake in glioblastoma cells and BBB permeability. All PDEVs displayed nanoscale sizes ranging from 103.5 to 141 nm with negative surface charge and a spherical morphological shape observed via SEM. The drug release kinetics was assessed using different mathematical models depicting the PDEVs prolonged drug release with < 50% releasing over 21 days. The cytotoxicity studies showed that the PDEVs resulted in a higher cell viability in the non-cancerous cell line compared to A172 glioblastoma cell line. The cellular internalization of the drug showed poor uptake of blank PDEVs compared to loaded PDEVs in glioblastoma cells. The BBB permeability test showed that ginger and aloe EVs permeated the BBB whilst BCS blank and loaded EVs did not permeate the BBB. This delivery system improves the ability of plant-derived extracellular vesicles to cross the blood-brain barrier, addressing a key challenge in delivering treatments to the brain. Through successful encapsulation of itraconazole, it paves the way for glioblastoma treatment by repurposing itraconazole with improved efficacy and reduced side effects. Furthermore, this can be incorporated in various drug delivery vehicles depending on the route of administration and therapeutic outcome i.e. intranasal, intravenous, or oral route. Future studies focus on determining the composition of PDEVs to enable engineering strategies for next generation targeting via surface modification.

|
Scooped by
mhryu@live.com
January 3, 4:36 PM
|
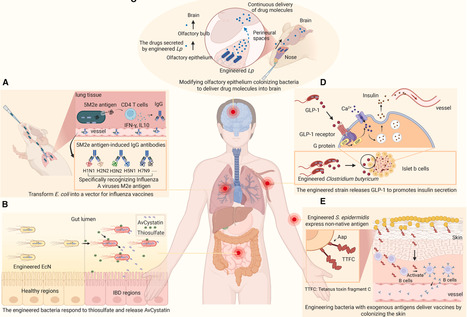
Engineered bacterial systems have been widely utilized as versatile biotherapeutic platforms in disease intervention, yet their therapeutic efficacy remains limited. Recent advancements in synthetic biology have enabled the systematic deconstruction of bacteria through rational computational modeling and modular-genetic circuit design, thereby overcoming intrinsic functional limitations. These engineered bacteria represent a promising therapeutic strategy characterized by enhanced spatiotemporal precision, biosafety profiles, and scalable production. Moreover, the advent of sophisticated genetic engineering technologies has facilitated the expansion of bacterial therapeutic applications into new areas, including vaccine development and tumor-microenvironment modulation. Here, we systematically examine recent advances in the development and testing of engineered bacteria across different organs on the basis of tissue specificity. We further summarize the therapeutic roles of engineered bacteria in tumor treatment and metabolic-disorder management. Additionally, we outline advanced technologies for engineered bacterial development, emphasizing their critical roles in optimizing the overall clinical efficacy of therapeutic agents after system-level optimization.

|
Scooped by
mhryu@live.com
January 3, 4:17 PM
|
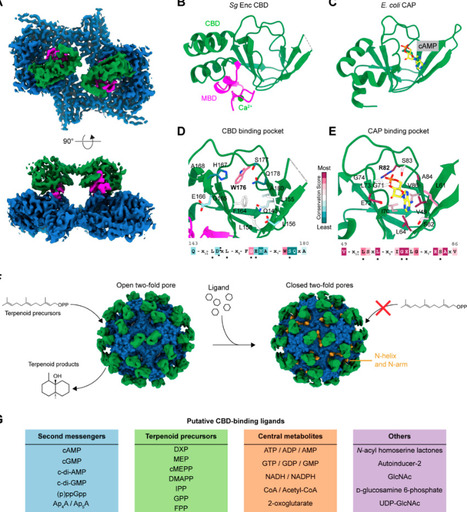
Encapsulins are self-assembling protein nanocompartments widely distributed across prokaryotes that sequester diverse enzymes. While most encapsulin systems studied thus far are involved in nutrient storage or oxidative stress response, recent bioinformatic and experimental studies have also demonstrated their involvement in secondary metabolism, particularly terpenoid biosynthesis. In this perspective, we first present a comprehensive analysis of Family 2B encapsulin gene clusters likely involved in terpene or terpenoid biosynthetic pathways. We then highlight the structural features of Family 2B encapsulin shells, with a focus on their pore properties and putative ligand-binding domains. We review the mechanisms of enzyme cargo loading in Family 2B systems and examine known examples of terpenoid synthesis compartmentalized within Family 2B encapsulin shells. This is followed by a discussion of the molecular logic and potential functional advantages of enzyme encapsulation. Finally, we consider outstanding questions and future research directions aimed at elucidating the molecular details and physiological implications of encapsulin-mediated bacterial terpene biosynthesis.
|
 Your new post is loading...
Your new post is loading...

























database