 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 2:22 PM
|
Adenosine-to-inosine (A-to-I) RNA editing by ADAR enzymes shapes transcript fate and underpins emerging RNA editing therapeutics, yet predicting which adenosines are edited remains difficult. We introduce ADAR-GPT, a model-agnostic fine-tuning framework that adapts a GPT-class language model to classify editing at candidate sites using sequence context in standardized 201 nt windows with the target adenosine explicitly marked. We train and evaluate on GTEx liver data (n=131 samples) at a clinically relevant 15% editing threshold, using a two-stage continual fine-tuning approach where lower thresholds serve as curriculum data to progressively sharpen decision boundaries. Using sequence data, ADAR-GPT demonstrates competitive or superior performance when benchmarked against established computational approaches, including convolutional and foundation model architectures, achieving a better balance of recall, precision, and specificity alongside stronger operating-curve metrics. The approach is reproducible and portable across GPT backbones without architectural changes. Beyond accurate site classification, ADAR-GPT provides practical adenosine scoring to prioritize experimental targets and inform guide RNA design, with a framework adaptable to new datasets and model architectures.

|
Scooped by
mhryu@live.com
Today, 1:40 PM
|
Microbes play a pivotal role in the Earth’s carbon cycle, regulating greenhouse gas fluxes by emitting, fixing and transforming CO2. Among them, acetogens stand out for their ability to fix CO2 through the Wood–Ljungdahl pathway, an ancient, highly energy-efficient route to acetyl-CoA that operates at thermodynamic limits. By coupling hydrogen (H2) or carbon monoxide oxidation to CO2 fixation, acetogens conserve energy while generating biomass and valuable products such as ethanol and acetate. These features position them as promising microbial cell factories for sustainable bioproduction via gas fermentation. Recent advances in metabolic engineering and synthetic biology have expanded the production spectrum of acetogens, enabling production of platform chemicals at lab-to-commercial scale. Yet, CO2-only bioconversion remains energetically challenging compared to syngas-based applications, requiring innovative solutions in strain development, bioprocess optimisation and integration of renewable energy sources. This review highlights the central role of model acetogens in anaerobic CO2 conversion, covering their metabolic capabilities, strain development and emerging bioprocess strategies to unlock their potential for low-carbon biomanufacturing.

|
Scooped by
mhryu@live.com
Today, 1:20 PM
|
Designing proteins that bind with high affinity to hydrophilic protein target sites remains a challenging problem. Here we show that RFdiffusion can be conditioned to generate protein scaffolds that form geometrically matched extended β-sheets with target protein edge β-strands in which polar groups on the target are complemented with hydrogen bonding groups on the design. We use this approach to design binders against edge-strand target sites on KIT, PDGFRɑ, ALK-2, ALK-3, FCRL5, NRP1, and α-CTX, and obtain higher (pM to mid nM) affinities and success rates than unconditioned RFdiffusion. Despite sharing β-strand interactions, designs have high specificity, reflecting the precise customization of interacting β-strand geometry and additional designed binder-target interactions. A binder-KIT co-crystal structure is nearly identical to the design model, confirming the accuracy of the design approach. The ability to robustly generate binders to the hydrophilic interaction surfaces of exposed β-strands considerably increases the range of computational binder design. This study demonstrates the capability of deep learning protein design models in generating functionally validated β-strand pairing interfaces, expanding the structural diversity of de novo binding proteins and accessible target surfaces.

|
Scooped by
mhryu@live.com
Today, 12:50 PM
|
The rapid growth of biodiesel production generates large amounts of crude glycerol, a low-value byproduct with environmental and economic challenges. Here, we present an engineered yeast coculture system combining Saccharomyces cerevisiae and Yarrowia lipolytica to convert crude glycerol into isopropanol, a liquid organic hydrogen carrier. The system integrates metabolic engineering, cell surface display pairing, immobilization, and continuous cultivation in fibrous bed bioreactors. In S. cerevisiae, glycerol use was improved by transporter optimization, pathway redirection, and flux shift from ethanol to isopropanol. In Y. lipolytica, ethanol from S. cerevisiae was redirected to isopropanol by acetyl-CoA reinforcement, malonyl-CoA diversion, and NADPH availability. Optimized pairing and inoculation ratios enhanced stability and yield. The consortia achieved complete glycerol utilization and three reuse cycles over 180 h. With pure glycerol, 28.34 g/l isopropanol was produced, while crude glycerol reached 86.06% of this yield. This strategy offers a scalable, modular route to convert biodiesel byproducts into bioenergy carriers.

|
Scooped by
mhryu@live.com
January 9, 5:06 PM
|
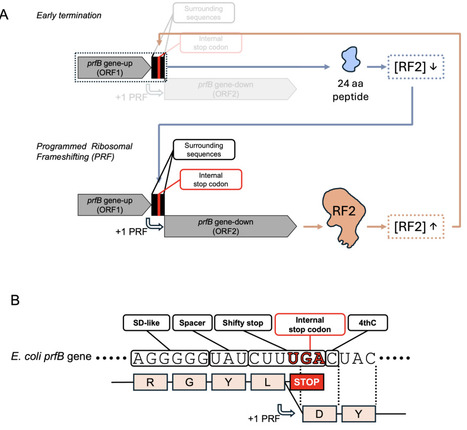
Understanding why some traits are maintained whereas others are repeatedly lost is a central question in evolutionary biology. Here, we address this problem through an analysis of the evolutionary dynamics of autoregulation of the prfB, which encodes peptide-chain release factor 2 (RF2), a key factor in bacterial translation termination. RF2 recognizes UGA and UAA stop codons and catalyzes the release of the completed polypeptide. In many species, prfB contains an internal UGA stop codon, triggering premature translation termination by RF2 itself. Complete RF2 translation depends on a +1 programmed ribosomal frameshifting (PRF) event on the internal stop codon, which occurs more frequently when RF2 levels are low, resulting in autoregulation of prfB expression. While widespread, this autoregulatory mechanism has been lost in multiple bacterial lineages. We combined phylogenetics, experimental evolution and molecular genetics to investigate the evolutionary forces behind this loss. We found no significant correlation between PRF loss and stop codon usage using phylogenetically informed analyses, and PRF disruption in Pseudomonas fluorescens SBW25 had no detectable fitness effect. However, engineered mutations that reduced frameshifting caused fitness defects, which were compensated by two classes of mutation: (i) mutations that impair specific ribosomal proteins, and (ii) single-nucleotide deletions in prfB that adjust the reading frame and bypass the internal stop codon. These results suggest that compensatory mutations facilitate the loss of prfB autoregulation under RF2-limiting conditions. We discuss three potential scenarios that could account for this process.

|
Scooped by
mhryu@live.com
January 9, 4:45 PM
|
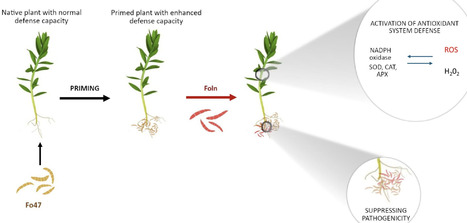
Plants rely on specialized adaptive mechanisms to enhance resistance against environmental stress. One such mechanism, priming, enables faster and stronger defence responses upon subsequent stress exposure. This study examines whether the non-pathogenic Fusarium oxysporum Fo47 primes flax by colonizing roots and activating antioxidant defences. Flax plants primed with Fo47 and those treated with both Fo47 and the pathogenic strain F. oxysporum Foln were analysed for fungal colonization, PR genes expression and antioxidant systems: enzymatic (ROS metabolism-related genes expression, catalase and superoxide dismutase activity, hydrogen peroxide and superoxide anion levels) and non-enzymatic (phenolic compound content and antioxidant potential). The results demonstrate that Fo47 colonises host tissues, significantly reducing Foln penetration and colonisation, particularly in primed plants. Root-specific suppression of Foln by Fo47 was stronger than systemic suppression in shoots. Fo47 induced early chitinase and NADPH oxidases D transcript accumulation and reduced superoxide anion level in roots, likely triggering defence activation. Notably, Fo47 also activated both enzymatic and non-enzymatic antioxidant systems in shoots, suggesting a systemic priming effect. These findings underscore the potential of non-pathogenic F. oxysporum strains in sustainable plant protection strategies.

|
Scooped by
mhryu@live.com
January 9, 4:29 PM
|
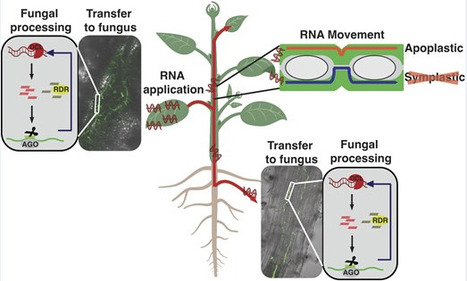
Foliar application of double-stranded RNA (dsRNA) as RNA interference (RNAi)-based bio-pesticides represents a sustainable alternative to chemical-based crop protection strategies. A key feature of RNAi in plants is its ability to act non-cell autonomously, a process that plays a critical role in plant development and protection against pathogens. Whether RNAi induced by foliar dsRNA application acts non-cell autonomously remains debated, with the mechanisms and implications of this movement largely unexplored. We show that upon foliar application, dsRNA enters the leaf vasculature and moves to vegetative, reproductive, and belowground tissues in multiple plant species. Unprocessed mobile dsRNA was detected in the apoplast, being maintained in new growth, indicating apoplastic rather than symplastic transport. Mobile dsRNA could transfer to infecting fungi, where it was processed and loaded by the fungal RNAi machinery to elicit gene silencing. Using a novel biochemical purification technique and small RNA sequencing, we detected functional small interfering RNA species derived from foliar-applied dsRNA that elicit effective silencing in both the applied and distal tissue types. Our mechanistic dissection of the uptake and movement of dsRNA provides crucial insights into the mode of action of RNAi biopesticides and stands to add significant benefit to this emerging field of plant protection.

|
Scooped by
mhryu@live.com
January 9, 4:21 PM
|
Bayesian optimization has become widely popular across various experimental sciences due to its favorable attributes: it can handle noisy data, perform well with relatively small data sets, and provide adaptive suggestions for sequential experimentation. While still in its infancy, Bayesian optimization has recently gained traction in bioprocess engineering. However, experimentation with biological systems is highly complex and the resulting experimental uncertainty requires specific extensions to classical Bayesian optimization. Moreover, current literature often targets readers with a strong statistical background, limiting its accessibility for practitioners. In light of these developments, this review has two aims: first, to provide an intuitive and practical introduction to Bayesian optimization; and second, to outline promising application areas and open algorithmic challenges, thereby highlighting opportunities for future research in machine learning.

|
Scooped by
mhryu@live.com
January 9, 3:57 PM
|
The composition of the vaginal microenvironment has significant implications for gynecologic and obstetric outcomes. Where a Lactobacillus-dominated microenvironment is considered optimal, a polymicrobial environment is associated with increased risk for female reproductive diseases. Recent work examined bacteria-derived extracellular vesicles (bEVs) as an important mode of microbe-host communication that may influence reproductive outcomes. However, in order to communicate with female reproductive tissues, bEVs must penetrate the protective cervicovaginal mucus barrier. We demonstrate increased diffusion of bEVs compared to whole bacteria. Additionally, we evaluate the uptake of bEVs by, and the resulting effects on, human vaginal epithelial, endometrial, and placental cells, highlighting potential mechanisms of action by which vaginal dysbiosis contributes to gynecologic and obstetric diseases. Taken together, our work demonstrates the ability of bEVs to mediate female reproductive outcomes and highlights their potential as therapeutic modalities for treating dysbiosis and dysbiosis-associated diseases in the female reproductive tract.

|
Scooped by
mhryu@live.com
January 9, 3:31 PM
|
RNA capped with dinucleoside polyphosphates has been discovered in bacteria and eukaryotes only recently. The likely mechanism of this specific capping involves direct incorporation of dinucleoside polyphosphates by RNA polymerase as noncanonical initiating nucleotides. However, how these compounds bind into the active site of RNA polymerase during transcription initiation is unknown. Here, we explored transcription initiation in vitro, using a series of DNA templates in combination with dinucleoside polyphosphates and model RNA polymerase from Thermus thermophilus. We observed that the transcription start site can vary on the basis of the compatibility of the specific template and dinucleoside polyphosphate. Cryo-electron microscopy structures of transcription initiation complexes with dinucleoside polyphosphates revealed that both nucleobase moieties can pair with the DNA template. The first encoded nucleotide pairs in a canonical Watson–Crick manner, whereas the second nucleobase pairs noncanonically in a reverse Watson–Crick manner. Our work provides a structural explanation of how dinucleoside polyphosphates initiate RNA transcription. Using cryo-electron microscopy technologies, Serianni and Škerlová et al. reveal how NpnNs initiate bacterial transcription as noncanonical RNA caps by showing one nucleobase pairing with the template in canonical mode while the other pairs in reverse Watson–Crick mode.

|
Scooped by
mhryu@live.com
January 9, 3:21 PM
|
Knowledge of bacterial flagella has largely come from studies of the simpler motors of Escherichia coli and Salmonella enterica. However, many bacteria harbor more complex motors. The function, mechanisms and evolution associated with such auxiliary motor structures are unclear. Here we deploy structural, genetic, biochemical and functional approaches to characterize complex adaptations of the flagellar motor in Campylobacter jejuni. We observed an E ring formed by 17 FlgY homodimers around the MS ring, a cage-like structure made of FcpMNO and PflD, and PflA–PflB interactions in a spoke–rim formation between the E ring and cage. These scaffolds stabilized the 17 torque-generating stator complexes. Phylogenetic analyses suggest an ancient origin and widespread prevalence of the E ring and spokes across diverse flagellated bacteria, and co-option of type IV pilus components in the ancestral motor of phylum Campylobacterota. Collectively, these data provide insight into the assembly, function and evolution of complex flagellar motors. Structural, genetic, functional and biochemical analyses of the complex flagellar motor of Campylobacter jejuni reveal structural adaptations with an ancient origin also found more widely across bacterial species, including elements exapted from the type IV pilus machinery.

|
Scooped by
mhryu@live.com
January 9, 3:07 PM
|
Sustainable bioenergy is pivotal for the global transition from fossil fuels to a circular bioeconomy, yet conventional biomass conversion is hindered by limitations in efficiency, cost, and versatility. This review examines how contemporary interdisciplinary advances are overcoming these challenges. We survey the convergence of synthetic biology, genomics, artificial intelligence (AI), and chemistry, which is revitalizing bioenergy production through the engineering of optimized biomass. Key strategies range from genomic editing of energy crops for enhanced nutrient efficiency and tailored lignin content to the development of AI-informed smart biorefineries. As a prime exemplar of this synergy, we present an in-depth case study on autoluminescent plants. This frontier application harnesses the fungal bioluminescence pathway (FBP) to directly convert photosynthetic energy into sustainable visible light. The FBP's unique reliance on the endogenous metabolite caffeic acid establishes a transformative platform for autonomous biological illumination. Our analysis underscores that an integrated approach, spanning omics, engineering, and agronomy, is critical for solving complex bioengineering problems and realizing the vision of high-brightness plants. We conclude by proposing that the next paradigm shift will be driven by generative AI, transitioning research and development from subject-specific inquiries to a holistic model of multidisciplinary convergence, thereby accelerating the realization of advanced, sustainable plant-based energy.

|
Scooped by
mhryu@live.com
January 9, 11:59 AM
|
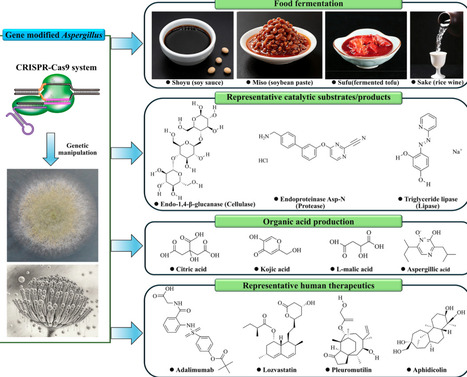
The genus Aspergillus comprises over 600 species of filamentous fungi. This genus significantly impacts human health, food fermentation, and industrial biotechnology. With the in-depth research and applications of Aspergillus species in many fields, the establishment of efficient gene editing technologies is crucial for functional genomics studies and cell factory development. The clustered regularly interspaced short palindromic repeats and associated protein (CRISPR-Cas9) system, as a newly developed and powerful genome editing tool, has demonstrated exceptional potential for precise genetic modifications in various Aspergillus species. The continuous advancement of CRISPR-Cas9 technology has enabled precise gene editing and modification in both pathogenic and industrial Aspergillus strains, thereby driving innovations in pathogenicity attenuation, metabolic engineering, and functional genomics. Therefore, this review provides a concise overview of the CRISPR-Cas9 system, detailing its composition, working mechanism, and key functional features such as the role of the Cas9 protein and the protospacer adjacent motifs (PAMs). Subsequently, we focus on the transformative applications of CRISPR-Cas9 in Aspergillus species, discussing its pivotal roles in elucidating pathogenic mechanisms, disrupting mycotoxin biosynthesis, and employing metabolic engineering to enhance the production of industrial enzymes, organic acids, and valuable natural products. Finally, we discuss future challenges and promising opportunities for applying CRISPR-Cas9 technology to advance the industrial biotechnology of Aspergillus species.
|

|
Scooped by
mhryu@live.com
Today, 2:13 PM
|
The apoplast is an important battlefield in plant–pathogen interactions. The late blight oomycete pathogen Phytophthora infestans, for instance, secretes cystatin-like protease inhibitors EpiC1 and EpiC2B to suppress C14, a papain-like immune protease secreted by tomato. Here, we found that P. infestans also secretes two distinct papain-like proteases termed Pain1 and Pain2, which are transcriptionally induced during infection. Both Pains promote P. infestans infection, but not when their catalytic residues are mutated. Strikingly, EpiC1 and EpiC2B preferentially inhibit tomato C14 rather than self-produced Pains, suggesting that they coevolved with Pains to avoid self-inhibition. To mimic the avoidance of inhibition by EpiCs, we engineered C14 (eC14) with seven Pain1 residues that potentially disturb the EpiCs–C14 interface. This eC14 is less sensitive to inhibition by EpiCs and enhances resistance to P. infestans infection. This strategy demonstrates that a pathogen-inspired protein engineering approach can increase crop resistance to plant pathogens.

|
Scooped by
mhryu@live.com
Today, 1:24 PM
|
Rapid and precise detection of small-molecule metabolites is crucial for optimizing the bioproduction processes. Cell-free systems (CFSs) offer an ideal platform for developing such biosensors due to their speed and suitability for automation. However, transcription factor (TF)-based biosensors, which are key elements for metabolite sensing, suffer from a severe bottleneck in in vitro environments. Their performance is often compromised due to the absence of nucleoid-associated proteins and different DNA topology compared to in vivo conditions. Here, we present a systematic framework for rationally engineering high-performance TF biosensors optimized for CFSs through integrated control of TF availability and promoter sequence. Using an itaconate (ITA)-responsive biosensor regulated by the LysR-type transcriptional regulator ItcR as a model system, we demonstrate that modulating TF supply and redesigning promoter elements substantially enhance sensitivity and dynamic range. Promoter dissection revealed that upstream sequences that function normally in vivo interfered with regulated transcription in CFSs, and that truncation to remove this region restored inducible behavior. Subsequent fine-tuning of the −35 and −10 motifs enhanced RNA polymerase recruitment and regulator interaction, resulting in 19-fold higher maximum signal, a 3.3-fold lower detection limit (0.003 g/L ITA), and a steeper dose-response curve (Hill slope increase from 2.7 to 34.7). The same promoter engineering strategy also improved a 3-hydroxypropionate-responsive biosensor, demonstrating its generality across distinct TF-promoter systems. Collectively, this framework establishes a rational, modular approach for constructing high-performance, topology-aware biosensors in CFSs, directly enabling high-throughput screening and automated biofoundry integration for synthetic biology and metabolic engineering applications.

|
Scooped by
mhryu@live.com
Today, 1:12 PM
|
The presence or absence of some genes in a genome can influence whether other genes are likely to be present or absent. Understanding these gene co-occurrence and avoidance patterns reveals fundamental principles of genome organization, with applications ranging from evolutionary reconstruction to rational design of synthetic genomes. PanForest, presented here, uses random forest classifiers to predict the presence and absence of genes in genomes from the set of other genes present. Performance statistics output by PanForest reveal how predictable each gene’s presence or absence is, based on the presence or absence of other genes in the genome. Further, PanForest produces statistics indicating the importance of each gene in predicting the presence or absence of each other gene. The PanForest software can run serially or in parallel, thereby facilitating the analysis of pangenomes at Network of Life scale. A pangenome of 12,741 accessory genes in 1,000 Escherichia coli genomes was analyzed in around 5 hours using 8 processors. To demonstrate PanForest’s utility, we present a case study and show that certain genes associated with resistance to antimicrobial drugs reliably predict the presence or absence of other genes associated with resistance to the same drug. Further, we highlight several associations between those genes and others not known to be associated with antimicrobial resistance (AMR), or associated with resistance to other drugs. We envisage PanForest’s use in studies from multiple disciplines concerning the dynamics of gene distributions in pangenomes ranging from biomedical science and synthetic biology to molecular ecology.

|
Scooped by
mhryu@live.com
Today, 12:44 PM
|
The growing imperative to ensure food safety, preserve ecological integrity, and mitigate public health risks associated with pesticide residues has driven a critical demand for highly sensitive optical sensors. In this regard, optical biosensors, including fluorescence (FL), colorimetry (CL), surface-enhanced Raman scattering (SERS), surface plasmon resonance (SPR), and chemiluminescence strategies, have been developed for pesticide detection. This review aims to provide a comprehensive summary of both fundamental knowledge and advancements in the field of optical biosensors for pesticide detection. The advantages of these biosensors are highlighted, such as excellent sensitivity, high specificity, and on-site application. Subsequently, a detailed overview of the sensing mechanism of optical biosensors based on different molecular recognition elements [e.g., enzymes, antibodies, aptamers, molecularly imprinted polymers (MIPs), and supramolecular host–guest complexes] is provided. Finally, perspectives are offered on the current challenges and future directions of pesticide biosensors. This review is expected to serve as a fundamental guide for researchers in the field of optical biosensors for pesticide detection and to provide insights and avenues to enhance the performance of existing sensing mechanisms in applications across diverse fields.

|
Scooped by
mhryu@live.com
January 9, 4:55 PM
|
Transcription factors regulate gene expression with DNA-binding domains (DBDs) and activation domains. Despite evidence to the contrary, DBDs are often assumed to be the primary mediators of transcription factor (TF) interactions with DNA and chromatin. Here, we used fast single-molecule tracking of transcription factors in living cells to show that short activation domains can control the fraction of molecules bound to chromatin. Stronger activation domains have higher bound fractions and longer residence times on chromatin. Furthermore, mutations that increase activation domain strength also increase chromatin binding. This trend was consistent in four different activation domains and their mutants. This effect further held for activation domains appended to three different structural classes of DBDs. Stronger activation domains with high chromatin-bound fractions also exhibited increased binding to the p300 coactivator in proximity-assisted photoactivation experiments. Genome-wide measurements indicate these activation domains primarily control the occupancy of binding rather than the genomic location. Taken together, these results demonstrate that very short activation domains play a major role in tethering transcription factors to chromatin.

|
Scooped by
mhryu@live.com
January 9, 4:34 PM
|
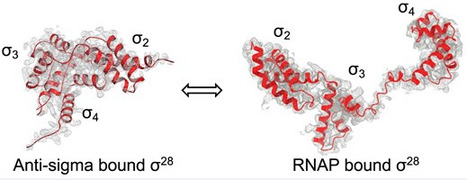
Late flagellar genes in Pseudomonas aeruginosa are transcribed by the group 3 sigma factor, FliA (σ28). σ28 drives the expression of flagellin, which assembles into the flagellar filament in this monoflagellated bacterium. This function is suppressed by the anti-sigma factor FlgM. Here, we present the 1.95Å resolution crystal structure of σ28–FlgM complex, along with a 3.4Å structure of σ28RNAP open promoter complex determined using single particle cryo-electron microscopy from P. aeruginosa. The σ28 adopts a compact conformation upon binding to the anti-sigma factor FlgM, which contacts all three domains of the sigma factor. This conformation is neither conducive to interactions with RNA polymerase nor the promoter DNA. The cryo-EM structure reveals base-specific interactions of σ28 domain 4 (σ4) with −35 element, flipping of −11 base of the template strand, novel interactions of template strand with domain 2 (σ2) and 3 (σ3), and partial insertion of sigma finger into the active site cleft, offering unique features of group 3 sigma interactions with promoter DNA. Perturbation of key residues affects transcription in vitro and flagellar phenotypes as well as bacterial motility in vivo. Analysis of the structural data presented here reveals new insights into transcription regulation of late flagellar genes.

|
Scooped by
mhryu@live.com
January 9, 4:25 PM
|
The trans cleavage activity of Cas12a has been extensively used for the detection of biomolecules. Different Cas12a orthologues exhibit faster or slower trans cleavage kinetics, making some orthologues more suited for sensitive molecular detection. Ionic strength of reaction buffers and mutations that change the electrostatic environment near the RuvC active site have also been reported to strongly influence trans cleavage kinetics. Studying three commonly used Cas12a orthologues (FnCas12a, AsCas12a, and LbCas12a), we report that electrostatic interactions near the RuvC active site are critical for their trans cleavage activity. Alanine substitution of arginine and lysine residues in the Nuc domain can abolish trans cleavage while modestly reducing cis cleavage. Substitutions in the RuvC lid and substitutions to introduce positively charged residues in the Nuc could enhance both cis and trans cleavage. These Cas12a variants improved DNA detection and genome editing efficacy. Overall, this study provides a blueprint for rationally engineering the DNase activities of Cas12a.

|
Scooped by
mhryu@live.com
January 9, 4:05 PM
|
Urinary tract infections represent one of the most prevalent bacterial diseases, yet current diagnostic and research methodologies are hampered by inadequate culture media that fail to replicate the bladder biochemical environment. Conventional artificial urine formulations contain undefined components, lack essential nutrients, or inadequately support urinary microbiome (urobiome) growth. To address these limitations, we developed SimUrine, a fully defined synthetic urine medium that aims to replicate human bladder chemistry while supporting diverse microbial growth requirements. SimUrine was systematically developed through iterative optimization of multi-purpose artificial urine, incorporating defined concentrations of carbon sources, vitamins, trace elements, and amino acids within physiologically relevant ranges. The modular design enables component substitution without complete reformulation, facilitating customization for culturomics, antimicrobial susceptibility testing, and microbial ecology studies, while reducing batch-to-batch variability associated with authentic urine. Performance evaluation demonstrated SimUrine's capability to support the growth of fastidious urobiome members, including Lactobacillus species, Aerococcus urinae, and Corynebacterium riegelii, which fail to proliferate in conventional minimal media. Physicochemical characterization confirmed that SimUrine formulation exhibits properties within normal human urine ranges for density, conductivity, osmolarity, and viscosity, ensuring physiological relevance. Clinical applications revealed reduced antibiotic susceptibility compared to standard media, suggesting a more accurate representation of in vivo conditions. Co-culture experiments using E. coli and Enterococcus faecalis demonstrated previously unobserved microbial interactions, highlighting SimUrine's utility for investigating urobiome dynamics. SimUrine represents a significant advancement in urobiome research methodology, providing a standardized, reproducible platform for investigating the urobiome under physiologically relevant conditions, potentially improving fundamental understanding and clinical diagnostic approaches.

|
Scooped by
mhryu@live.com
January 9, 3:37 PM
|
Engineering synthetic intrinsically disordered proteins (synIDPs) enables regulation of biomolecular condensation and protein solubility. However, limited understanding of how sequence-dependent interaction cooperativity relates to the fitness impacts of synIDPs on endogenous cellular processes constrains our design capability. Here, to circumvent this design challenge, we present a systematic directed evolution method for the evolution of synIDPs capable of mediating diverse phase behaviors in living cells. The selection methods allow us to evolve a toolbox of synIDPs with distinct phase behaviors and thermoresponsive features in living cells, leading to the evolution of synthetic condensates. The reverse-selection method further allows us to select synIDPs as solubility tags. We demonstrate the applications of the evolved synIDPs in protein circuits to (1) regulate intracellular protein activity and (2) reverse antibiotic resistance. Our systematic evolution and selection strategies provide a versatile platform for developing synIDPs for broad applications in synthetic biology and biotechnology. Ma et al. developed a directed evolution method with various selection strategies for the evolution of synthetic intrinsically disordered proteins capable of forming condensates with desired properties in living cells.

|
Scooped by
mhryu@live.com
January 9, 3:24 PM
|
Biomanufacturing provides a more sustainable alternative to fossil-based chemical manufacturing. 3-Hydroxypropionic acid (3HP) is a top Department of Energy value-added chemical and precursor to bio-plastics, yet cost-effective microbial production remains elusive. Here, we establish the acid-tolerant yeast Issatchenkia orientalis as a robust host for low-pH 3HP biosynthesis. Genome-scale modeling identifies the β-alanine pathway as optimal, offering the highest theoretical yield and lowest oxygen requirement. Thermodynamic analysis confirms its favorability under acidic conditions. Using sequence similarity network analysis, we discover highly active aspartate 1-decarboxylase (PAND), β-alanine-pyruvate aminotransferase (BAPAT), and 3HP dehydrogenase (YDFG), which significantly improve the pathway efficiency. Next, to further elevate the production, pathway optimization through multi-copy PAND integration, byproduct elimination (knockouts of pyruvate decarboxylase and glycerol-3-phosphate dehydrogenase), and reinforcement of aspartate flux by overexpression of pyruvate carboxylase and aspartate amino transferase improves the titer to 29 g/L in shake flasks. Fed-batch fermentation at pH 4 with low-cost corn steep liquor medium further increases the production to 92 g/L with 0.7 g/g yield and 0.55 g/L/h productivity. Techno-economic analysis indicates that such performance could potentially enable a financially viable process for sustainable acrylic acid production. This work establishes I. orientalis as a next-generation platform for cost-effective 3HP production and paves the way toward industrial commercialization. 3-Hydroxypropionic acid (3HP) is a top Department of Energy value-added chemical and precursor to bioplastics, yet cost-effective microbial bioproduction remains elusive. Here the authors establish efficient 3HP production in an acid tolerant yeast and validate its financially viability.

|
Scooped by
mhryu@live.com
January 9, 3:16 PM
|
While the degradation of plastics into molecules and micro- and nanoplastics (MNPs) through abiotic processes is increasingly well documented, bacterial-driven biodegradation remains poorly understood. Yet, as microbial strategies to mitigate plastic pollution gain traction, identifying the full spectrum of degradation products, from soluble molecules to microplastics and nanoplastics, is essential for elucidating both toxicological impacts and fundamental degradation mechanisms. In this study, we developed an integrated analytical method combining thermodesorption and pyrolysis gas chromatography–mass spectrometry (TDS/Py-GC-MS). A tailored liquid–liquid extraction protocol was optimized for both polymers and applied to culture media containing microbial communities in contact with solid plastic samples. This approach enabled the identification of specific depolymerization products from polystyrene (PS) and polyvinyl chloride (PVC), including known markers (e.g., phthalic anhydride, 2-ethylhexan-1-ol, acenaphthene) and novel aromatic compounds likely associated with microbial activity, such as (1E)-1-benzylideneindene, 2-ethylcyclopentan-1-one, and benzene in samples from superworms intestinal microbiota cultivated with PS or PVC film at different temperature (n = 1). In terms of fragmentation, PS particles were recovered in varying quantities, while no PVC fragments were detected. This study provides a robust analytical framework to characterize plastic degradation and identify molecular markers of degradation.

|
Scooped by
mhryu@live.com
January 9, 12:02 PM
|
Pulmonary infections caused by Aspergillus fumigatus are a global health concern. In the lungs, A. fumigatus employs a variety of virulence and adaptation mechanisms that allows it to survive and evade local immune responses. A. fumigatus protein kinases co-ordinately regulate adaptation to the lung environment and antifungal agents. However, to improve our understanding of the role of these fungal protein kinases in subverting the antifungal responses exhibited by specific host cells, in vitro infection models should be developed. Here, we describe a functional genomics approach to evaluate the role of A. fumigatus protein kinases upon infection of airway epithelial cells, leveraging the use of our existing library of A. fumigatus deletion mutants for kinase functions and bar-seq. We infected A549 alveolar epithelial cells with a pool of 111 A. fumigatus barcoded null mutant strains in genes encoding protein kinases and used bar-seq to identify which of these are involved in fungal survival upon A549 infection. To assess the optimal method for understanding host-pathogen interactions, three approaches aimed at removing extracellular conidia were evaluated: pharmacological treatment with the antifungal nystatin, washes of unbound conidia, and differential fluorescent staining of extracellular and intracellular conidia followed by cell sorting. The identified candidates were further validated using targeted approaches. Our data revealed new roles for A. fumigatus protein kinases during infection of airway epithelial cells. Modification of the experimental setup allowed us to differentiate between the genes associated with uptake and those required to withstand other stresses imposed by exposure to mammalian cells. This protocol provides a new platform to ascribe pathogenicity-associated functions to A. fumigatus genes and allows prioritization for downstream experiments in mice, potentially reducing the number of studies needed in live animals. This is particularly pertinent, as genome-wide barcoded collections with thousands of strains have become available.
|
 Your new post is loading...
Your new post is loading...






























Stephanopoulos