Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
November 28, 2012 2:56 AM
|
Custom assembly of TAL effector repeat array (transcription activator–like effector nuclease, TALEN and TALE transcription factor, TALE-TF) Services: Design and assemby of TAL effector repeat arrays including TALE transcription factors (TALE-TFs) and nucleases (TALENs)Clone into an expression vector of your choice. Related Services: Produce 5′-capped mRNA of TALEN pairs or TALE-TF. in vitro translation to generate functional TALEN proteins in vitro cleavage assay to assess TALEN activity in vitro transcription and capping to generate capped poly(A) mRNA in vivo cleavage assay

|
Suggested by
Eli
November 15, 2012 2:56 AM
|
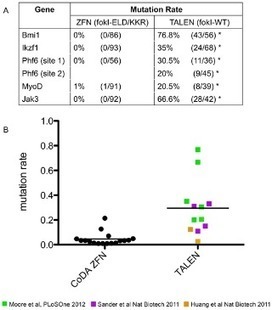
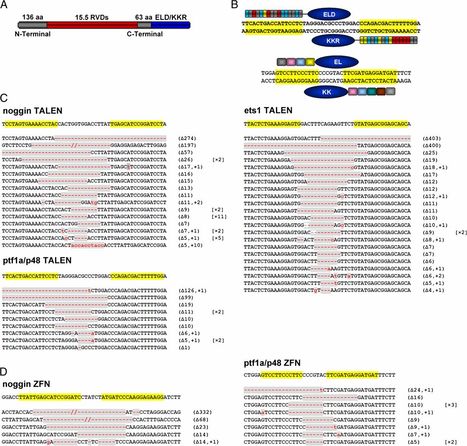
Here, we assess the relative efficiencies of these technologies for inducing somatic DNA mutations in mosaic zebrafish. We find that TALENs exhibited a higher success rate for obtaining active nucleases capable of inducing mutations than compared with Context-Dependent Assembly (CoDA) ZFNs. For example, all six TALENs tested induced DNA mutations at genomic target sites while only a subset of CoDA ZFNs exhibited detectable rates of mutagenesis. TALENs also exhibited higher mutation rates than CoDA ZFNs that had not been pre-screened using a bacterial two-hybrid assay, with DNA mutation rates ranging from 20%–76.8% compared to 1.1%–3.3%. Furthermore, the broader targeting range of TALENs enabled us to induce mutations at the methionine translation start site, sequences that were not targetable using the CoDA ZFN platform. TALENs exhibited similar toxicity to CoDA ZFNs, with >50% of injected animals surviving to 3 days of life. Taken together, our results suggest that TALEN technology provides a robust alternative to CoDA ZFNs for inducing targeted gene-inactivation in zebrafish, making it a preferred technology for creating targeted knockout mutants in zebrafish.
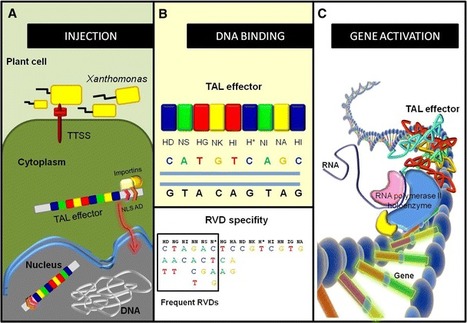
via @NicoDnce Muñoz Bodnar et al. (2012) Pathogenic bacteria of the Xanthomonas and Ralstonia genus have developed resourceful strategies creating a favorable environment to multiply and colonize their host plants. One of these strategies involves the secretion and translocation of several families of effector proteins into the host cell. The transcription activator-like effector (TALE) family forms a subset of proteins involved in the direct modulation of host gene expression. TALEs include a number of tandem 34-amino acid repeats in their central part, where specific residues variable in two adjacent positions determine DNA-binding in the host genome. The specificity of this binding and its predictable nature make TALEs a revolutionary tool for gene editing, functional analysis, modification of target gene expression, and directed mutagenesis. Several examples have been reported in higher organisms as diverse as plants, Drosophila, zebrafish, mouse, and even human cells. Here, we summarize the functions of TALEs in their natural context and the biotechnological perspectives of their use.
Via Nicolas Denancé

|
Scooped by
dromius
November 7, 2012 5:32 PM
|
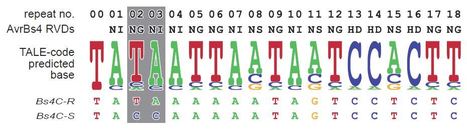
via Tom Schreiber (get well soon! and thanks) Transcription activator-like effector (TALE) proteins of the plant pathogenic bacterial genus Xanthomonas bind to and transcriptionally activate host susceptibility genes, promoting disease. Plant immune systems have taken advantage of this mechanism by evolving TALE binding sites upstream of resistance (R) genes. For example, the pepper Bs3 and rice Xa27 genes are hypersensitive reaction plant R genes that are transcriptionally activated by corresponding TALEs. Both R genes have a hallmark expression pattern in which their transcripts are detectable only in the presence and not the absence of the corresponding TALE. By transcriptome profiling using next-generation sequencing (RNA-seq), we tested whether we could avoid laborious positional cloning for the isolation of TALE-induced R genes. In a proof-of-principle experiment, RNA-seq was used to identify a candidate for Bs4C, an R gene from pepper that mediates recognition of the Xanthomonas TALE protein AvrBs4. We identified one major Bs4C candidate transcript by RNA-seq that was expressed exclusively in the presence of AvrBs4. Complementation studies confirmed that the candidate corresponds to the Bs4C gene and that an AvrBs4 binding site in the Bs4C promoter directs its transcriptional activation. Comparison of Bs4C with a nonfunctional allele that is unable to recognize AvrBs4 revealed a 2-bp polymorphism within the TALE binding site of the Bs4C promoter. Bs4C encodes a structurally unique R protein and Bs4C-like genes that are present in many solanaceous genomes seem to be as tightly regulated as pepper Bs4C. These findings demonstrate that TALE-specific R genes can be cloned from large-genome crops with a highly efficient RNA-seq approach.

|
Suggested by
Eli
November 5, 2012 2:50 AM
|
via @eli88fine Transcription activator-like effector nucleases (TALENs) are an approach for directed gene disruption and have been proved to be effective in various animal models. Here, we report that TALENs can induce somatic mutations in Xenopus embryos with reliably high efficiency and that such mutations are heritable through germ-line transmission. We modified the Golden Gate method for TALEN assembly to make the product suitable for RNA transcription and microinjection into Xenopus embryos. Eight pairs of TALENs were constructed to target eight Xenopus genes, and all resulted in indel mutations with high efficiencies of up to 95.7% at the targeted loci. Furthermore, mutations induced by TALENs were highly efficiently passed through the germ line to F1 frogs. Together with simple and reliable PCR-based approaches for detecting TALEN-induced mutations, our results indicate that TALENs are an effective tool for targeted gene editing/knockout in Xenopus.

|
Suggested by
Eli
October 30, 2012 10:15 AM
|
via @eli88fine In the past decade, Xenopus tropicalis has emerged as a powerful new amphibian genetic model system, which offers all of the experimental advantages of its larger cousin, Xenopus laevis. Here we investigated the efficiency of transcription activator-like effector nucleases (TALENs) for generating targeted mutations in endogenous genes in X. tropicalis. For our analysis we targeted the tyrosinase (oculocutaneous albinism IA) (tyr) gene, which is required for the production of skin pigments, such as melanin. We injected mRNA encoding TALENs targeting the first exon of the tyr gene into two-cell-stage embryos. Surprisingly, we found that over 90% of the founder animals developed either partial or full albinism, suggesting that the TALENs induced bi-allelic mutations in the tyr gene at very high frequency in the F0 animals. Furthermore, mutations tyr gene were efficiently transmitted into the F1 progeny, as evidenced by the generation of albino offspring. These findings have far reaching implications in our quest to develop efficient reverse genetic approaches in this emerging amphibian model.

|
Scooped by
dromius
October 22, 2012 3:29 AM
|
Verdier et al (2012) Genomes of the rice (Oryza sativa) xylem and mesophyll pathogens Xanthomonas oryzae pv. oryzae (Xoo) and pv. oryzicola (Xoc) encode numerous secreted transcription factors called transcription activator-like (TAL) effectors. In a few studied rice varieties, some of these contribute to virulence by activating corresponding host susceptibility genes. Some activate disease resistance genes. The roles of X. oryzae TAL effectors in diverse rice backgrounds, however, are poorly understood. Xoo TAL effectors that promote infection by activating SWEET sucrose transporter genes were expressed in TAL effector-deficient X. oryzae strain X11-5A, and assessed in 21 rice varieties. Some were also tested in Xoc on variety Nipponbare. Several Xoc TAL effectors were tested in X11-5A on four rice varieties. Xoo TAL effectors enhanced X11-5A virulence on most varieties, but to varying extents depending on the effector and variety. SWEET genes were activated in all tested varieties, but increased virulence did not correlate with activation level. SWEET activators also enhanced Xoc virulence on Nipponbare. Xoc TAL effectors did not alter X11-5A virulence. SWEET-targeting TAL effectors contribute broadly and non-tissue-specifically to virulence in rice, and their function is affected by host differences besides target sequences. Further, the utility of X11-5A for characterizing individual TAL effectors in rice was established.

|
Scooped by
dromius
October 10, 2012 2:44 AM
|
VANSTON, III., Oct. 9, 2012 /PRNewswire-iReach/ -- (October 9, 2012) – The Two Blades Foundation (2Blades) announced today the completion of a non-exclusive license for the TAL code genome engineering technology to Mendel Biotechnology, Inc., a leading plant science company developing improved plant varieties and crop protection solutions for global agriculture.

|
Scooped by
dromius
October 1, 2012 3:21 AM
|
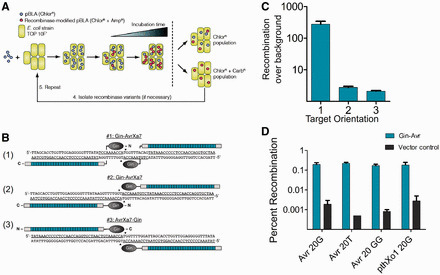
Mercer et al (2012) [...] Imperfect modularity with particular domains, lack of high-affinity binding to all DNA triplets, and difficulty in construction has hindered the widespread adoption of zinc-finger proteins (ZFPs) in unspecialized laboratories. The discovery of a novel type of DNA-binding domains in transcription activator-like effector (TALE) proteins from Xanthomonas provides an alternative to ZFPs. Here we describe chimeric TALE recombinases (TALERs): engineered fusions between a hyperactivated catalytic domain from the DNA invertase Gin and an optimized TALE architecture. We use a library of incrementally truncated TALE variants to identify TALER fusions that modify DNA with efficiency and specificity comparable to zinc-finger recombinases in bacterial cells. We also show that TALERs recombine DNA in mammalian cells. The TALER architecture described herein provides a platform for insertion of customized TALE domains, thus significantly expanding the targeting capacity of engineered recombinases and their potential applications in biotechnology and medicine.

|
Scooped by
dromius
September 28, 2012 3:14 AM
|
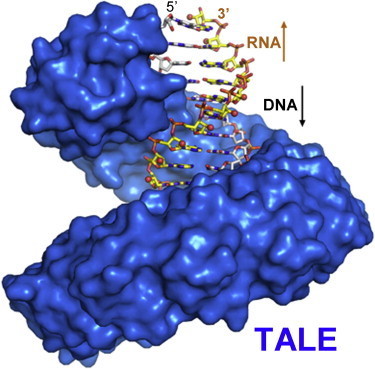
Yin et al (2012) via Tom Schreiber (thanks!) * TALE repeats specifically bind to DNA-RNA hybrids * The sequence of DNA determines binding specificity by TALE repeats * TALE repeats protect DNA-RNA hybrids from RNase H degradation * TALEs may be used to fight retroviral infection and modulate transcription Summary The transcription activator-like (TAL) effector targets specific host promoter through its central DNA-binding domain, which comprises multiple tandem repeats (TALE repeats). Recent structural analyses revealed that the TALE repeats form a superhelical structure that tracks along the forward strand of the DNA duplex. Here, we demonstrate that TALE repeats specifically recognize a DNA-RNA hybrid where the DNA strand determines the binding specificity. The crystal structure of a designed TALE in complex with the DNA-RNA hybrid was determined at a resolution of 2.5 Å. Although TALE repeats are in direct contact with only the DNA strand, the phosphodiester backbone of the RNA strand is inaccessible by macromolecules such as RNases. Consistent with this observation, sequence-specific recognition of an HIV-derived DNA-RNA hybrid by an engineered TALE efficiently blocked RNase H-mediated degradation of the RNA strand. Our study broadens the utility of TALE repeats and suggests potential applications in processes involving DNA replication and retroviral infections.

|
Scooped by
dromius
September 27, 2012 3:06 AM
|
(via Tom Schreiber [again! ... keep going]) Valton et al 2012, JBC, Cellectis, France Within the past two years, TAL DNA binding domains have emerged as the new generation of engineerable platform for production of custom DNA binding domains. However, their recently described sensitivity to cytosine methylation represents a major bottleneck for genome engineering applications. Using a combination of biochemical, structural and cellular approaches, we were able to identify the molecular basis of such sensitivity and propose a simple, drug-free and universal method to overcome it. Background: TALE-based technologies are poised to revolutionize the field of biotechnology however, their sensitivity to cytosine methylation may drastically restrict the range of their
applications.
Results: TALE repeat N* is able to proficiently bind to 5-methylated cytosine.
Conclusion: Sensitivity of TALE DNA binding domains to cytosine methylation can be overcome by using TALE repeat N*. Significance: Utilization of TALE repeat N* enables broadening the scope of TALE-based technologies.

|
Scooped by
dromius
September 25, 2012 3:25 AM
|
Heritable gene targeting in zebrafish using customized TALENs - Nature Biotech.
http://www.nature.com/nbt/journal/v29/n8/full/nbt.1939.html Nature Biotechnology29,699–700(2011) Studies of targeted gene modifications are of great interest in basic research as well as for clinical and agricultural applications1. In the February issue of Nature Biotechnology, two articles reported genomic modifications using transcription activator-like (TAL) effectors2, 3. Using fusion proteins, each comprising a TAL effector DNA binding domain and a FokI cleavage domain, Miller et al.2 reported that TAL effector nucleases (TALENs) successfully disrupted target genes in cultured human cells. Zhang et al.3 showed that TAL effectors can be used to regulate endogenous gene transcription. Compared with zinc-finger proteins4, 5, TAL effectors permit more predictable and specific binding to target DNA6, and therefore allow researchers to engineer genomes precisely without the need for laborious screening to identify a DNA binding domain with the requisite specificity. TALENs can induce DNA double-stranded breaks (DSBs) in yeast7. Gene targeting using TALENs has also been achieved in nematodes8 and human pluripotent cells9. However, it has not, to our knowledge, yet been demonstrated in a vertebrate organism. Here we report the use of TALENs to disrupt both of the two endogenous zebrafish genes we targeted and show that the mutations are transmitted through the germ line.

|
Scooped by
dromius
September 24, 2012 7:01 AM
|
suggested by Tom Schreiber (thanks!) Establishment of efficient genome editing tools is essential for fundamental research, genetic engineering, and gene therapy. Successful construction and application of transcription activator-like effector nucleases (TALENs) in several organisms herald an exciting new era for genome editing. We describe the production of two active TALENs and their successful application in the targeted mutagenesis of silkworm, Bombyx mori, whose genetic manipulation methods are parallel to those of Drosophila and other insects. We will also show that the simultaneous expression of two pairs of TALENs generates heritable large chromosomal deletion. Our results demonstrate that (i) TALENs can be used in silkworm and (ii) heritable large chromosomal deletions can be induced by two pairs of TALENs in whole organisms. The generation and the high frequency of TALENs-induced targeted mutagenesis in silkworm will promote the genetic modification of silkworm and other insect species.
|

|
Scooped by
dromius
November 15, 2012 3:02 AM
|
Maresca et al (2012) Custom designed nucleases (CDNs) greatly facilitate genetic engineering by generating a targeted DNA double-strand break (DSB) in the genome. Once a DSB is created, specific modifications can be introduced around the breakage site during its repair by two major DNA damage repair (DDR) mechanisms: the dominant but error-prone non-homologous end joining (NHEJ) pathway and the less-frequent but precise homologous recombination (HR) pathway. Here we describe ObLiGaRe, a new method for site-specific gene insertions which uses the efficient NHEJ pathway and acts independently of HR. This method is applicable with both zinc finger nucleases (ZFNs) and Tale nucleases (TALENs) and has enabled us to insert a 15 kb inducible gene expression cassette at a defined locus in human cell lines. In addition, our experiments have revealed a previously underestimated error-free nature of NHEJ and provided new tools to further characterize this pathway under physiological and pathological conditions.

|
Suggested by
Eli
November 15, 2012 2:53 AM
|
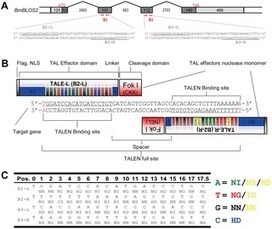
Engineered nucleases are proteins that are able to cleave DNA at specified sites in the genome. These proteins have recently been used for gene targeting in a number of organisms. We showed earlier that zinc finger nucleases (ZFNs) can be used for generating gene-specific mutations in Bombyx mori by an error-prone DNA repair process of non-homologous end joining (NHEJ). Here we test the utility of another type of chimeric nuclease based on bacterial TAL effector proteins in order to induce targeted mutations in silkworm DNA. We designed three TAL effector nucleases (TALENs) against the genomic locus BmBLOS2, previously targeted by ZFNs. All three TALENs were able to induce mutations in silkworm germline cells suggesting a higher success rate of this type of chimeric enzyme. The efficiency of two of the tested TALENs was slightly higher than of the successful ZFN used previously. Simple design, high frequency of candidate targeting sites and comparable efficiency of induction of NHEJ mutations make TALENs an important alternative to ZFNs.

|
Scooped by
dromius
November 9, 2012 2:54 AM
|
via Matt Moscou, thanks! With a rapidly growing fan base, gene-editing tools enable experiments from basic biomedical research to crop science. Researchers use these tools to edit genomes at sites of their choosing. This article provides an overview on current applications, providers and ways to generate genome editing tools, specifically nucleases.

|
Suggested by
Przemyslaw Bidzinski
November 6, 2012 3:20 AM
|
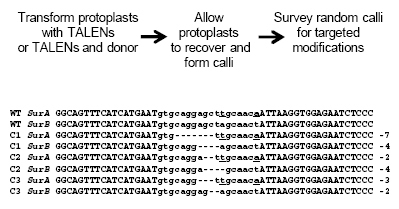
Thanks Przemek! Zhang et al. (2012) The ability to precisely engineer plant genomes offers much potential for advancing basic and applied plant biology. Here we describe methods for the targeted modification of plant genomes using transcription activator like effector nucleases (TALENs). Methods were optimized using Nicotiana tabacum (tobacco) protoplasts and TALENs targeting the acetolactate synthase (ALS) gene. Optimal TALEN scaffolds were identified using a protoplast-based single-strand annealing (SSA) assay in which TALEN cleavage creates a functional YFP gene, enabling quantification of TALEN activity by flow cytometry. SSA activity data for TALENs with different scaffolds correlated highly with their activity at endogenous targets, as measured by high throughput DNA sequencing of PCR products encompassing the TALEN recognition sites. TALENs made with optimized scaffolds introduced targeted mutations in ALS in 30% of transformed cells, and frequencies of targeted gene insertion approximated 14%. These efficiencies made it possible to recover genome modifications without selection or enrichment regimes: 31.5% of tobacco calli generated from protoplasts transformed with TALEN-encoding constructs had TALEN-induced mutations in ALS, and of 16 calli characterized in detail, all had mutations in one allele each of the duplicate ALS genes (SurA and SurB). In calli derived from cells treated with a TALEN and a 322 bp donor molecule differing by 6 bp from the ALS coding sequence, 3.5% showed evidence of targeted gene replacement. The optimized reagents described and implemented in plant protoplasts should be useful for targeted modification of cells from diverse plant species and using a variety of means for reagent delivery.

|
Scooped by
dromius
October 30, 2012 3:42 PM
|
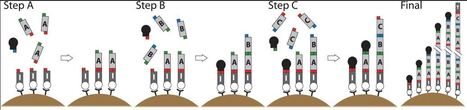
via Jens Boch (thanks!) DNA built from modular repeats presents a challenge for gene synthesis. We present a solid surface-based sequential ligation approach, which we refer to as iterative capped assembly (ICA), that adds DNA repeat monomers individually to a growing chain while using hairpin ‘capping’ oligonucleotides to block incompletely extended chains, greatly increasing the frequency of full-length final products. Applying ICA to a model problem, construction of custom transcription activator-like effector nucleases (TALENs) for genome engineering, we demonstrate efficient synthesis of TALE DNA-binding domains up to 21 monomers long and their ligation into a nuclease-carrying backbone vector all within 3 h. We used ICA to synthesize 20 TALENs of varying DNA target site length and tested their ability to stimulate gene editing by a donor oligonucleotide in human cells. All the TALENS show activity, with the ones >15 monomers long tending to work best. Since ICA builds full-length constructs from individual monomers rather than large exhaustive libraries of pre-fabricated oligomers, it will be trivial to incorporate future modified TALE monomers with improved or expanded function or to synthesize other types of repeat-modular DNA where the diversity of possible monomers makes exhaustive oligomer libraries impractical.

|
Scooped by
dromius
October 29, 2012 6:42 PM
|
movie: http://vimeo.com/49902809 Introduction You don't know what TAL effectors actually are? We reviewed the recent literature for you, to give you a quick overview of this exciting new field of research. Golden Gate Standard Assembling multiple gene constructs in frame without leaving scars is not possible with existing iGEM standards. We therefore introduce the new Golden-Gate Standard that is fully compatible with RFC10. The TAL Vector Targeting a sequence and not doing something to it, is like throwing mechanics at your car. Your car will not get any better only the mechanics will get mad. Because we know this, we bring the tools you need to actually work with DNA.To make it even more easy these tools are deliverd already inside the final TAL backbone, just add the sequence and you're ready. GATE Assembly Kit We have invented a super-fast, super-easy and super-cheap Method for custom TAL effector construction. Learn about the theory behind the TAL effector toolkit, how we created it and why we choose this design. Using the Toolkit Our overall goal is to empower future iGEM teams to use the most exciting new technology synthetic biology has to offer. We therefore not only invented the GATE assembly platform but wrote a step by step manual for super-easy custom TALE construction The Future of TAL Until now, almost three years after deciphering the TALE code, only two types of TAL Effectors have been developed: TALENs and TAL-TFs. We herein propose additional classes of TAL effectors. Experiments and Results We not only rigorously tested if our in vitro TALE gene assembly method works but also if our TALE constructs actually work in a human cell line. Check out test design and results.

|
Scooped by
dromius
October 11, 2012 3:49 AM
|
Stratech Scientific offer SBI’s TAL Effector Assembly Service. Transcription Activator-Like Effectors can be used for genome editing and gene modulation. For TALENs, two engineered TAL effectors are designed that bind the targeted DNA sequence on either side of the chosen cut site in the genome. Errors in DNA repair at the site of cleavage lead to deletions and insertions, and permanent alteration of the targeted locus. Transcription Activator-Like Effector Activators or Repressors are engineered transcription factors created by fusing the TAL effector DNA binding domain that targets a specific promoter region to a transcription activation or repression protein domain. Receive ready-to-transfect TALE constructs under the control of your preferred promoter (CMV, EF-1α, or MSCV). [comment: no advert, just to reflect the variety of services]

|
Scooped by
dromius
October 2, 2012 2:55 AM
|
Carlson et al (2012 [via Tom Schreiber] Transcription activator-like effector nucleases (TALENs) are programmable nucleases that join FokI endonuclease with the modular DNA-binding domain of TALEs. Although zinc-finger nucleases enable a variety of genome modifications, their application to genetic engineering of livestock has been slowed by technical limitations of embryo-injection, culture of primary cells, and difficulty in producing reliable reagents with a limited budget. In contrast, we found that TALENs could easily be manufactured and that over half (23/36, 64%) demonstrate high activity in primary cells. Cytoplasmic injections of TALEN mRNAs into livestock zygotes were capable of inducing gene KO in up to 75% of embryos analyzed, a portion of which harbored biallelic modification. We also developed a simple transposon coselection strategy for TALEN-mediated gene modification in primary fibroblasts that enabled both enrichment for modified cells and efficient isolation of modified colonies. Coselection after treatment with a single TALEN-pair enabled isolation of colonies with mono- and biallelic modification in up to 54% and 17% of colonies, respectively. Coselection after treatment with two TALEN-pairs directed against the same chromosome enabled the isolation of colonies harboring large chromosomal deletions and inversions (10% and 4% of colonies, respectively). TALEN-modified Ossabaw swine fetal fibroblasts were effective nuclear donors for cloning, resulting in the creation of miniature swine containing mono- and biallelic mutations of the LDL receptor gene as models of familial hypercholesterolemia. TALENs thus appear to represent a highly facile platform for the modification of livestock genomes for both biomedical and agricultural applications.

|
Scooped by
dromius
September 28, 2012 3:23 AM
|
Coupling endonucleases with DNA end–processing enzymes to drive gene disruption - Nature Methods
Certo et al, Nature Methods9,973–975(2012) Targeted DNA double-strand breaks introduced by rare-cleaving designer endonucleases can be harnessed for gene disruption applications by engaging mutagenic nonhomologous end-joining DNA repair pathways. However, endonuclease-mediated DNA breaks are often subject to precise repair, which limits the efficiency of targeted genome editing. To address this issue, we coupled designer endonucleases to DNA end–processing enzymes to drive mutagenic break resolution, achieving up to 25-fold enhancements in gene disruption rates.

|
Scooped by
dromius
September 27, 2012 2:09 PM
|
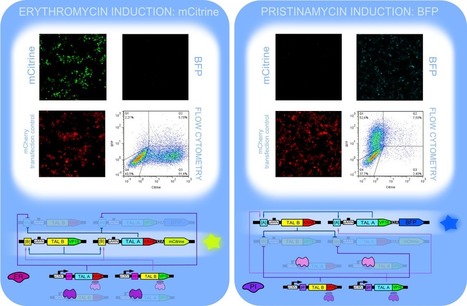
We designed an upgraded bistable genetic toggle switch based on orthogonal TAL repressors and activators which is composed of a pair of mutual repressors and a pair of activators extending the classical toggle swith with positive feedback loops. Simulations of the positive feedback loop switch demonstrated bistability even at low or no cooperativity. Stochastic and deterministic simulations indicate higher robustness in comparison to the mutual repressor switch. We experimentally tested the switch by monitoring production of two fluorescent protein reporters. We confirmed a clear bimodal distribution of reporter fluorescence and demonstrated adoption of stable states by induction with corresponding inducer molecules. The switch persisted in a stable state after the removal of inducer molecules, which confirmed the epigenitc bistability of our system.

|
Scooped by
dromius
September 25, 2012 3:26 AM
|
In vivo genome editing using a high-efficiency TALEN system - Nature
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11537.html Bedell et al. (2012) Nature The zebrafish (Danio rerio) is increasingly being used to study basic vertebrate biology and human disease with a rich array of in vivo genetic and molecular tools. However, the inability to readily modify the genome in a targeted fashion has been a bottleneck in the field. Here we show that improvements in artificial transcription activator-like effector nucleases (TALENs) provide a powerful new approach for targeted zebrafish genome editing and functional genomic applications1, 2, 3, 4, 5. Using the GoldyTALEN modified scaffold and zebrafish delivery system, we show that this enhanced TALEN toolkit has a high efficiency in inducing locus-specific DNA breaks in somatic and germline tissues. At some loci, this efficacy approaches 100%, including biallelic conversion in somatic tissues that mimics phenotypes seen using morpholino-based targeted gene knockdowns6. With this updated TALEN system, we successfully used single-stranded DNA oligonucleotides to precisely modify sequences at predefined locations in the zebrafish genome through homology-directed repair, including the introduction of a custom-designed EcoRV site and a modified loxP (mloxP) sequence into somatic tissue in vivo. We further show successful germline transmission of both EcoRV and mloxP engineered chromosomes. This combined approach offers the potential to model genetic variation as well as to generate targeted conditional alleles.

|
Scooped by
dromius
September 25, 2012 3:21 AM
|
Christian et al, (2012) suggested Tom Schreiber, thanks! The DNA binding domain of Transcription Activator-Like (TAL) effectors can easily be engineered to have new DNA sequence specificities. Consequently, engineered TAL effector proteins have become important reagents for manipulating genomes in vivo. DNA binding by TAL effectors is mediated by arrays of 34 amino acid repeats. In each repeat, one of two amino acids (repeat variable di-residues, RVDs) contacts a base in the DNA target. RVDs with specificity for C, T and A have been described; however, among RVDs that target G, the RVD NN also binds A, and NK is rare among naturally occurring TAL effectors. Here we show that TAL effector nucleases (TALENs) made with NK to specify G have less activity than their NN-containing counterparts: fourteen of fifteen TALEN pairs made with NN showed more activity in a yeast recombination assay than otherwise identical TALENs made with NK. Activity was assayed for three of these TALEN pairs in human cells, and the results paralleled the yeast data. The in vivo data is explained by in vitro measurements of binding affinity demonstrating that NK-containing TAL effectors have less affinity for targets with G than their NN-containing counterparts. On targets for which G was substituted with A, higher G-specificity was observed for NK-containing TALENs. TALENs with different N- and C-terminal truncations were also tested on targets that differed in the length of the spacer between the two TALEN binding sites. TALENs with C-termini of either 63 or 231 amino acids after the repeat array cleaved targets across a broad range of spacer lengths – from 14 to 33 bp. TALENs with only 18 aa after the repeat array, however, showed a clear optimum for spacers of 13 to 16 bp. The data presented here provide useful guidelines for increasing the specificity and activity of engineered TAL effector proteins.
|



 Your new post is loading...
Your new post is loading...