Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
October 30, 3:12 PM
|
Rai and Wang 2025
Citrus canker caused by Xanthomonas citri subsp. citri (Xcc) is an important citrus disease worldwide. PthA4 is the most important pathogenicity gene of Xcc and encodes a transcription activator-like effector (TALE) secreted by the type III secretion system. PthA4 is known to activate the expression of CsLOB1, the canker susceptibility gene and a transcription factor, to cause citrus canker symptoms. Extensive effort was made to identify downstream targets of CsLOB1 to investigate the mechanism underlying canker symptom development. However, none of the identified CsLOB1 target genes has been confirmed to be involved in citrus canker development. Here, we first identified the direct targets of CsLOB1 by generating a promoter-uidA (GUS) reporter fusion construct for the 13 genes highly induced by both PthA4 and CsLOB1 and monitored the reporter activity in Nicotiana benthamiana leaves co-expressing CsLOB1. Agrobacterium tumefaciens-mediated transient expression of CsLOB1 activated seven gene promoters in N. benthamiana: Cs7g18460, Orange1.1t00600, Cs6g17190, Cs7g32410 (CsEXP2), Cs2g27100, Cs2g20750 (CsEG1), and Cs9g17380. Next, we constructed designer TALEs (dTALEs) to target unique sequences in the promoters of the seven direct target genes of CsLOB1 and transformed them into the XccpthA4::Tn5 mutant. Our results indicated that a combination of five and seven dTALEs caused canker-like symptoms in the inoculated citrus leaves. In addition, dTALECsEXP2 and dTALECsEG1 caused water soaking and pustules, which are typical canker symptoms. Taken together, Xcc indirectly activates CsEXP2 and CsEG1 via PthA4-CsLOB1 to cause canker symptoms. Identification of direct targets of CsLOB1 provides alternative targets for genetic improvement of citrus against canker via genome editing. Copyright © 2025 The Author(s). This is an open access article distributed under the CC BY-NC-ND 4.0 International license.

|
Scooped by
dromius
October 17, 2:46 AM
|
Transient expression of a transcription activator-like effector (TALE)-based epigenetic editor demonstrates durable therapeutic silencing in non-human primates.

|
Scooped by
dromius
August 23, 5:11 AM
|
Sohini Deb et al. 2025
Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight of rice, translocates multiple transcription activator-like effectors (TALEs) into rice cells. The TALEs localize to the host cell nucleus, where they bind to the DNA in a sequence-specific manner and enhance gene expression to promote disease susceptibility. Xoo strain PXO99A encodes 19 TALEs, but the host targets of all these TALEs have not been defined. A meta-analysis of rice transcriptome profiles revealed a gene annotated as flavonol synthase/flavanone-3 hydroxylase (henceforth OsS5H/FNS-03g) to be highly induced upon Xoo infection. Further analyses revealed that this gene is induced by PXO99A using Tal9b, a broadly conserved TALE of Xoo. Disruption of tal9b rendered PXO99A less virulent. OsS5H/FNS-03g functionally complemented its Arabidopsis homologue AtDMR6, a well-studied disease susceptibility locus. Biochemical analyses suggested that OsS5H/FNS-03g is a bifunctional protein with salicylic acid 5′ hydroxylase (S5H) and flavone synthase-I (FNS-I) activities. Further, an exogenous application of apigenin, the flavone that is enzymatically produced by OsS5H/FNS-03g, on rice leaves promoted virulence of PXO99A tal9b–. Overall, our study suggests that OsS5H/FNS-03g is a bifunctional enzyme, and its product, apigenin, is potentially involved in promoting Xoo virulence. Copyright © 2025 The Author(s). This is an open access article distributed under the CC BY-NC-ND 4.0 International license.

|
Scooped by
dromius
March 26, 3:58 AM
|
(via T. Schreiber, thx) Fan et al. 2025
Transcription activator-like effector-linked deaminases (TALEDs) use their single-stranded DNA (ssDNA)-specific adenosine deaminase TadA8e to mediate A-to-G editing in mitochondrial DNA (mtDNA). The working mechanism of this process is unknown, hindering the development of more effective TALEDs. Here we reveal that TALED-mediated A-to-G editing relies on the formation of an ssDNA region through base excision repair (BER), which is triggered by double-stranded DNA-specific cytidine deaminase (DddA)-induced C-to-U deamination. We develop a series of enhanced TALEDs (eTALED6s) with increased editing efficiency by replacing DddA with the high-activity variant DddA6 and fusing human uracil DNA glycosylase to TadA8e. By further engineering TadA8e, the resulting eTALED6Rs induces efficient on-target editing with reduced bystander editing and off-target editing at the DNA and RNA levels. Lastly, we use eTALED6 and eTALED6R to install a pathogenic mutation in mtDNA. Revealing the mechanism of TALED-mediated A-to-G editing demonstrates that enhancing BER increases editing efficiency.

|
Scooped by
dromius
March 11, 3:33 PM
|
(via T. Schreiber, thx) Jia et al. 2025
Transcription activator-like effectors (TALEs) secreted from Xanthomonas oryzae pv oryzae (Xoo) function as a pathogenicity factor to activate rice bacterial blight (BB) susceptibility, conforming to the gene-for-gene paradigm as well as resistance. Xoo pathotypes generally harbor one to three major TALEs targeting OsSWEET genes to determine pathogenicity; conversely, the immunity events mediated by minor TALEs have not been taken seriously. Here, we demonstrated that lipid transfer protein encoding gene OsLTPL23 positively regulates rice resistance to Xoo pathotype PXO61, and TalAE73PXO61, a representative member of the most widely distributed TALE family in 135 Xoo isolates, transcriptionally activates OsLTPL23 expression. Further, TalAE73PXO61 is an avirulence protein, causing effector-triggered immunity in compatible rice-Xoo interaction. In addition, reactive oxygen species accumulation, nitrate uptake, and salicylic acid homeostasis are transcriptionally and physiologically associated with OsLTPL23-dependent BB resistance.

|
Scooped by
dromius
January 31, 6:17 AM
|
Syed Mashab Ali Shah et al. 2025
Bacterial blight of cotton (BBC) caused by Xanthomonas citri pv. malvacearum (Xcm) is an important and destructive disease affecting cotton plants. Transcription activator-like effectors (TALEs) released by the pathogen regulate cotton resistance to the susceptibility. In this study, we sequenced the whole genome of Xcm Xss-V2-18 and identified eight tal genes: seven on the plasmids and one on the chromosome. Deletion and complementation experiments of Xss-V2-18 tal genes demonstrated that Tal1b is required for full virulence on cotton. Transcriptome profiling coupled with TALE-binding element prediction revealed that Tal1b targets GhSWEET14A04/D04 and GhSWEET14D02 simultaneously. Expression analysis confirmed the independent inducibility of GhSWEET14A04/D04 and GhSWEET14D02 by Tal1b, whereas GhSWEET14A04/D04 is additionally targeted by Tal1. Moreover, β-glucuronidase and Xa10-mediated hypersensitive response assays indicated that the effector-binding element (EBEs) are required for the direct and specific activation of the candidate targets by Tal1 and Ta1b. These insights enhance our understanding of the underlying mechanisms of bacterial blight in cotton and might lead to improved resistance through EBEs disruption or a TALE-trap strategy.

|
Scooped by
dromius
January 31, 6:10 AM
|
Liu et al. 2025
TALEN-mediated plant genome editing has the advantages of high specificity, independence from epigenetic modification, lack of PAM limitation, and feasibility for organelle editing due to no need for RNA binding. However, the assembly of TALEN vectors was difficult and laborious, affecting the popularity of TALEN genome editing in plants. In this study, a novel TALEN-based plant genome editing tool, ZQTALEN, was developed. This system comprises a total of nine plasmids categorized into three types. The TALE repeat units are obtained through PCR utilizing the template vector as the amplification template. These units are then assembled sequentially: first into donor vectors to form entry vectors, and subsequently transferred to destination vectors, resulting in the final binary vector. Characterized by the optimization of codon usage, assembly method of TALE repeat array, and vector backbone components, ZQTALEN has the advantages of easy, flexible and efficient assembly and less repeated sequences in the final vector. Using the ZQTALEN system, a TALEN binary vector was successfully constructed to target the endogenous Nramp5 gene in rice, resulting in the high-frequency acquisition of rice mutants. The ZQTALEN system has provided a versatile tool for genetic research in plants.

|
Scooped by
dromius
January 11, 12:58 PM
|
Karki 2025
This article comments on:
Gaudin C, Preveaux A, Aubineau N, Le Goff D, Jacques M-A, Chen NWG. 2025. A dTALE approach demonstrates that induction of common bean OVATE Family Protein 7 promotes resistance to common bacterial blight. Journal of Experimental Botany 76, 607–620. https://doi.org/10.1093/jxb/erae433
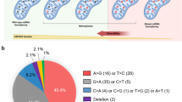
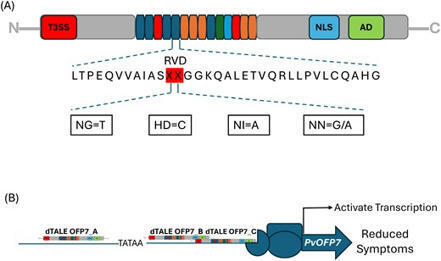
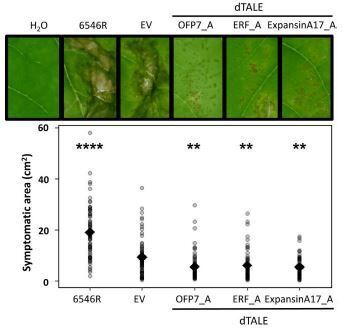
Bacteria belonging to the genus Xanthomonas impact >400 plant species worldwide. They are known to employ transcription activator-like effectors (TALEs) to modulate host gene expression, leading to disease. These TALEs can be designed to target specific DNA sequences which allows them to have broad applications in genome engineering. Gaudin et al. (2025) showcase the use of designer TALEs (dTALEs) to identify resistance genes against common bacterial blight of bean (CBB), a significant threat to bean production worldwide. Their findings identify three genes, PvOFP7, PvAP2-ERF1, and PvExpansinA17, as key for disease resistance, with PvOFP7 showing the highest differential expression, as well as significant reduction in disease symptoms and bacterial population when induced with dTALEs. This discovery opens up new perspectives on CBB resistance by linking PvOFP7-mediated defences to heat shock protein suppression and cell wall reinforcement, in addition to offering insights into the potential to use dTALEs in breeding resistant common bean varieties.

|
Scooped by
dromius
June 9, 2024 3:40 AM
|
Roeschlin et al. 2024
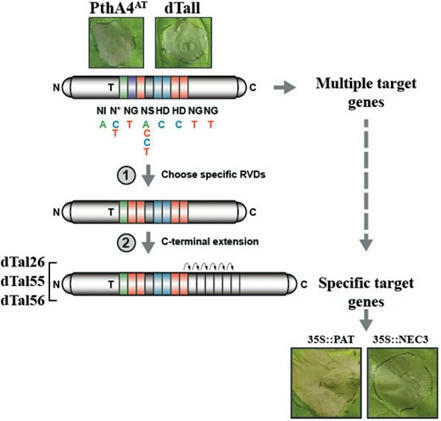
Transcription activator-like effectors (TALEs) in plant-pathogenic Xanthomonas bacteria activate expression of plant genes and support infection or cause a resistance response. PthA4AT is a TALE with a particularly short DNA-binding domain harboring only 7.5 repeats which triggers cell death in Nicotiana benthamiana; however, the genetic basis for this remains unknown. To identify possible target genes of PthA4AT that mediate cell death in N. benthamiana, we exploited the modularity of TALEs to stepwise enhance their specificity and reduce potential target sites. Substitutions of individual repeats suggested that PthA4AT-dependent cell death is sequence specific. Stepwise addition of repeats to the C-terminal or N-terminal end of the repeat region narrowed the sequence requirements in promoters of target genes. Transcriptome profiling and in silico target prediction allowed the isolation of two cell death inducer genes, which encode a patatin-like protein and a bifunctional monodehydroascorbate reductase/carbonic anhydrase protein. These two proteins are not linked to known TALE-dependent resistance genes. Our results show that the aberrant expression of different endogenous plant genes can cause a cell death reaction, which supports the hypothesis that TALE-dependent executor resistance genes can originate from various plant processes. Our strategy further demonstrates the use of TALEs to scan genomes for genes triggering cell death and other relevant phenotypes.

|
Scooped by
dromius
May 19, 2024 5:00 AM
|
Liu et al. 2024
The detection of DNA methylation at cytosine/guanine dinucleotide (CpG) islands in promoter regions of tumor suppressor genes has great potential for early cancer screening, diagnosis, and prognosis monitoring. Nevertheless, achieving accurate, sensitive, cost-effective, and quantitative detection of target methylated DNA remains challenging. Herein, we propose a novel piezoelectric sensor (series piezoelectric quartz crystal (SPQC)) based on transcription activator-like effectors (TALEs) for detecting DNA methylation of Ras association domain family 1 isoform A (RASSF1A) tumor suppressor genes (R-5mC). The sensor employs TALEs-Ni magnetic beads to specifically recognize and separate the R-5mC, thereby improving the detection selectivity. The TALEs-Ni magnetic beads-R-5mC complex is sheared by a nucleic acid enzyme (DNAzyme) to release the single-stranded DNA (ST). ST initiates a catalyzed hairpin assembly (CHA) reaction on the surface of the electrode, which in turn triggers the hybridization chain reaction (HCR) and silver staining for enhanced detection sensitivity. The strategy exhibits a linear response in the detection of R-5mC in the range of 1 fM to 1 nM with a detection limit of 0.79 fM. R-5mC as low as 0.01% can be detected, even in the presence of large numbers of unmethylated DNA. The detection of R-5mC in circulating cell-free DNA (cfDNA) derived from clinical plasma specimens of lung cancer patients yielded satisfactory results.

|
Scooped by
dromius
April 13, 2024 8:27 AM
|
BASF PLANT SCIENCE COMPANY GMBH
United States Patent Application 20240110193
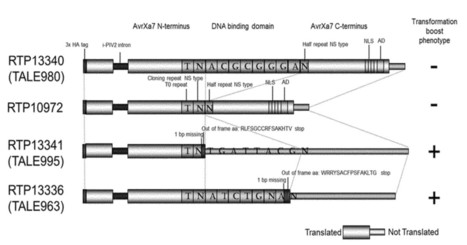
Although significant advances have been made in the field of transformation methods, a need continues to exist for improved methods to facilitate the ease, speed and efficiency of such methods for transformation of plants. Therefore, it was the objective of the present invention to provide an improved method having higher overall efficiency in the process of generation of transgenic plants. This objective is solved by the present invention. Surprisingly, it was shown in the studies underlying the present invention that introduction of a truncated transcription activator-like effector (TALE) polypeptide allows for a general improvement of transformation. The truncated TALE polypeptide comprises the N-terminus of a TALE polypeptide. In particular, the introduction allowed for an enhanced transformation efficacy, a faster growth of transformed calli, a faster generation of TO plants. Further, an increased biomass of generated TO plants was observed. Advantageously, in the T1 generation plants appeared to develop normally. The transformation efficacy was increased more than 2 fold over controls and plants were moved to the greenhouse 3 weeks earlier.
The observed effect was independent of nuclear localization and transcriptional activation as both corresponding domains are not functionally present in the truncated protein.

|
Scooped by
dromius
December 21, 2023 5:39 PM
|
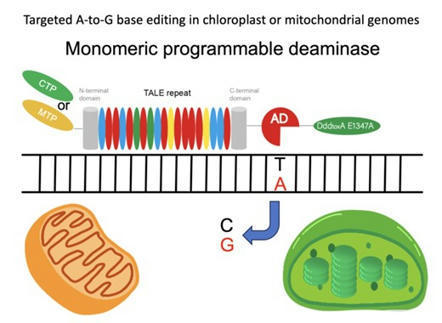
Zhou et al. 2023
Plastids and mitochondria are two intracellular organelles containing DNA encoding partial but essential components for their roles, photosynthesis and respiration. Precise base editing in both plastid and mitochondrial genomes would benefit their gene functional analysis and crop breeding. Targeted base editing in organellar genomes relies on a protein-based genome editing system that uses the TALE-DNA recognition motif with deaminases. This is because the efficient delivery of guide RNA for CRISPR/Cas9 systems into organelles is currently impossible. Since TALE-based base editors used in organellar genomes are usually dimeric types, here, we used targeted A-to-G base editing in Arabidopsis (Arabidopsis thaliana) plastid and mitochondrial genomes with monomeric TALE-based deaminase for easier assembling of vectors. As a result, inheritable targeted A-to-G base editing of ATPase subunit 6-2 (atp6-2) in plant mitochondrial genomes and of 16S ribosomal RNA (16S rRNA) in plastid genomes of Arabidopsis was successfully induced by monomeric TALE-based adenine deaminase without off-target mutations. The monomeric TALE-based adenine deaminases also demonstrated a preference for editing the 8th T on the same strand from the recognition end. Phenotypic analysis showed the A-to-G conversion at 1139A of plastid 16S rRNA conferred substantial spectinomycin resistance in Arabidopsis, but not the other two potential-resistant mutations at 1131T and 1137T, predicted from the previous bacterial data. Our study demonstrated the feasibility of monomeric TALE-based adenine deaminases in plant organelles and their potential contribution to the functional analyses of plant organelles with easier assembling.

|
Scooped by
dromius
October 23, 2023 6:30 PM
|
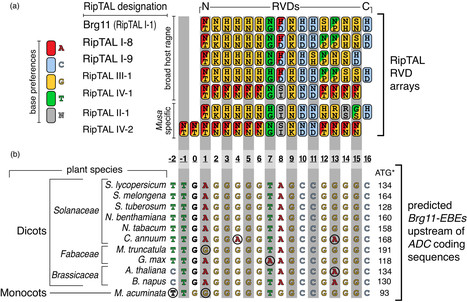
Gallas et al. 2023 Ralstonia solanacearum, a species complex of bacterial plant pathogens that causes bacterial wilt, comprises four phylotypes that evolved when a founder population was split during the continental drift ~180 million years ago. Each phylotype contains strains with RipTAL proteins structurally related to transcription activator-like (TAL) effectors from the bacterial pathogen Xanthomonas. RipTALs have evolved in geographically separated phylotypes and therefore differ in sequence and potentially functionality. Earlier work has shown that phylotype I RipTAL Brg11 targets a 17-nucleotide effector binding element (EBE) and transcriptionally activates the downstream arginine decarboxylase (ADC) gene. The predicted DNA binding preferences of Brg11 and RipTALs from other phylotypes are similar, suggesting that most, if not all, RipTALs target the Brg11-EBE motif and activate downstream ADC genes. Here we show that not only phylotype I RipTAL Brg11 but also RipTALs from other phylotypes activate host genes when preceded by the Brg11-EBE motif. Furthermore, we show that Brg11 and RipTALs from other phylotypes induce the same quantitative changes of ADC-dependent plant metabolites, suggesting that most, if not all, RipTALs induce functionally equivalent changes in host cells. Finally, we report transgenic tobacco lines in which the RipTAL-binding motif Brg11-EBE mediates RipTAL-dependent transcription of the executor-type resistance (R) gene Bs4C from pepper, thereby conferring resistance to RipTAL-delivering R. solanacearum strains. Our results suggest that cell death-inducing executor-type R genes, preceded by the RipTAL-binding motif Brg11-EBE, could be used to genetically engineer broad-spectrum bacterial wilt resistance in crop plants without any apparent fitness penalty.
|

|
Scooped by
dromius
October 24, 5:30 PM
|
Zaman et al. 2025
Secondary metabolites, including alkaloids, flavonoids, and tannins, are crucial for human health, agriculture, and ecosystem functioning. Their synthesis is often species-specific, influenced by both genetic and environmental factors. The increasing demand for these compounds across various industries highlights the need for advancements in plant breeding and biotechnological approaches. Transcription activator-like effector nucleases (TALENs) have emerged as a powerful tool for precise genome editing, offering significant potential for enhancing the synthesis of secondary metabolites in plants. However, while plant genome editing technologies have advanced significantly, the application of TALENs in improving secondary metabolite production and expanding genetic diversity remains underexplored. Therefore, this review aims to provide a comprehensive analysis of TALEN-mediated genome editing in plants, focusing on their role in enhancing secondary metabolite biosynthetic pathways and improving genetic diversity. The mechanisms underlying TALENs are examined, including their ability to target specific genes involved in the synthesis of bioactive compounds, highlighting comparisons with other genome editing tools such as CRISPR/Cas9. This review further highlights key applications in medicinal plants, particularly the modification of pathways responsible for alkaloids, flavonoids, terpenoids, and phenolic compounds. Furthermore, the role of TALENs in inducing genetic variation, improving stress tolerance, and facilitating hybridization in plant breeding programs is highlighted. Recent advances, challenges, and limitations associated with using TALENs for enhancing secondary metabolite production are critically evaluated. In this review, gaps in current research are identified, particularly regarding the integration of TALENs with multi-omics technologies and synthetic biology approaches. The findings suggest that while underutilized, TALENs offer sustainable strategies for producing high-value secondary metabolites in medicinal plants. Future research should focus on optimizing TALEN systems for commercial applications and integrating them with advanced biotechnological platforms to enhance the yield and resilience of medicinal plants.

|
Scooped by
dromius
September 10, 10:48 AM
|
Chen et al. 2025
Plant proteins that belong to the nonexpressor of pathogenesis-related (NPR) gene family are paralogous receptors of the plant defense hormone salicylic acid and essential regulators of hormone-dependent plant immunity against diseases caused by various pathogens. Previous studies have established NPR1 and NPR3 as a transcriptional activator and a transcriptional repressor, respectively, of defense-gene expression to promote and inhibit broad-spectrum resistance against different strains of pathogens. However, the regulatory mechanism that underlies the opposing roles of NPR1 and NPR3 in defense-gene activation remains unclear. Here, we report that a rice transcript splicing factor, Oryza sativa RNA-binding protein 11 (OsRBP11), promotes alternative splicing of OsNPR3 to modulate the defense function of OsNPR1 in rice plants infected by Xanthomonas oryzae pathovars, which are important bacterial pathogens of rice. We discovered that 11 transcription activator-like effectors identified in representative bacterial strains activate OsRBP11 expression. The OsRBP11 protein, in turn, facilitates alternative splicing of the OsNPR3 mRNA precursor, leading to the production of truncated OsNPR3 protein variants. The OsNPR3 variants exacerbate bacterial diseases by sequestering OsNPR1 from defense-gene activation. By contrast, both artificial and natural variations in OsRBP11 prevent the alternative splicing of OsNPR3, restore the defense function of OsNPR1, and enhance rice resistance to different bacterial strains. These findings not only reveal a novel regulatory pathway exploited by bacterial pathogens to facilitate their pathogenicity and subvert plant defense but also provide a genetic basis for biotechnological strategies aimed at developing broad-spectrum resistance in crops.

|
Scooped by
dromius
June 24, 12:38 PM
|
Im et al. 2025
Transcription activator-like effectors (TALEs) mimic eukaryotic transcriptional activators and translocate into host plant cells via the bacterial type III secretion system (T3SS) during pathogenic interactions. They play a crucial role in disease development by regulating host genes. Despite this, the regulatory mechanisms by which TALEs control OsWRKY transcription factors (TFs) remain poorly understood. In this study, we show that two TALEs from Xanthomonas oryzae pv. oryzae (Xoo) individually modulate two OsWRKY TFs, resulting in increased susceptibility and reduced host defense. Specifically, Xoo1219 and Xoo2145 activate the expression of OsWRKY104 and OsWRKY55, respectively, through direct interactions. OsWRKY104 increases the susceptibility to Xoo by activating OsSWEET11 and OsSWEET14, while OsWRKY55 suppresses host defense against Xoo by directly regulating OsWRKY62. These findings suggest that TALEs hijack the host's OsWRKY TFs to create a favorable environment for bacterial survival.

|
Scooped by
dromius
March 22, 8:15 AM
|
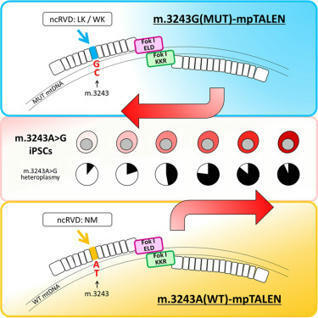
Yahata et al. 2025
Patient-derived induced pluripotent stem cells (iPSCs) are a useful pathological model for debilitating diseases caused by mitochondrial DNA (mtDNA) mutations. We established iPSCs derived from mitochondrial disease patients, heteroplasmic for the m.3243A>G mutation. The proportion of a selected mtDNA can be reduced by delivering a programmable nuclease into the mitochondria, and we developed various mtDNA-targeted Platinum TALENs (mpTALENs) to modify m.3243A>G-iPSC heteroplasmy levels in either wild-type or mutant direction. For TALEN optimization, the use of non-conventional repeat-variable di-residues (ncRVD)—LK/WK or NM—enhanced cleavage activity and specificity, and the replacement of conventional with obligate heterodimeric FokI nuclease domains increased target specificity and protected mtDNA from copy number depletion. In vitro, depending on whether wild-type or mutant mtDNA was targeted, we could obtain m.3243A>G-iPSCs with a higher or lower mutation load, while the cells retained their ability to differentiate into three germ layers. These results demonstrate that our mpTALEN optimization created a useful tool for altering heteroplasmy levels in m.3243A>G-iPSCs, improving the potential for studying mutation pathology. The enhanced efficiency also holds promise for using m.3243G(MUT)-mpTALEN as a therapeutic strategy for treating patients suffering from m.3243A>G mitochondrial diseases.

|
Scooped by
dromius
February 12, 5:19 PM
|
Yoshihisa et al. 2025 Key message Rice CC-BED NLR Xa1 recognizes TAL effectors through the interaction between ERF101 and TAL effectors. Abstract The rice Xa1 gene encodes a nucleotide-binding leucine-rich repeat receptor with an N-terminal coiled coil-zinc finger BED (CC-BED) domain. Xa1 recognizes the transcription activator-like (TAL) effectors of Xanthomonas oryzae pv. oryzae (Xoo) in the nucleus, triggering a number of immune responses, including hypersensitive cell death. We previously discovered that the rice transcription factor ERF101 directly interacts with Xa1, and functions as a positive regulator of Xa1-dependent immunity. However, the involvement of ERF101 in Xa1-induced immunity remains unclear. We herein demonstrated that the expression of the CC-BED domain in rice protoplasts inhibited Xa1-induced cell death. However, the CC-BEDC165A,C168A domain which has mutations of cysteine residues conserved in the zinc-finger motifs of BED domains and is essential for forming tetrahedral coordination geometry, failed to inhibit cell death or interact with ERF101. Therefore, Xa1-induced cell death appears to depend on the interaction between the BED domain and ERF101. In addition, we generated transgenic plants overexpressing N-terminal or C-terminal FLAG-tagged ERF101. FLAG-ERF101 transgenic plants exhibited reduced levels of Xa1-mediated immunity against Xoo, even though the overexpression of ERF101-FLAG or non-tagged ERF101 enhanced immunity. This result was consistent with the CC-BED domain interacting with C-terminal tagged ERF101, but not N-terminal tagged ERF101, whereas N-terminal and C-terminal tagged ERF101 both interacted with TAL effectors. Therefore, the interaction between the BED domain and ERF101 appears to be essential for the recognition of TAL effectors by Xa1.

|
Scooped by
dromius
January 31, 6:13 AM
|
Mormile et al. 2025
Bacterial transcription activator-like effectors (TALEs) promote pathogenicity by activating host susceptibility (S) genes. To understand the pathogenicity and host adaptation of Xanthomonas citri pv. malvacearum (Xcm), we assemble the genome and the TALE repertoire of three recent Xcm Texas isolates. A newly evolved TALE, Tal7b, activates GhSWEET14a and GhSWEET14b, different from GhSWEET10 targeted by a TALE in an early Xcm isolate. Activation of GhSWEET14a and GhSWEET14b results in water-soaked lesions. Transcriptome profiling coupled with TALE-binding element prediction identify a pectin lyase gene as an additional Tal7b target, quantitatively contributing to Xcm virulence alongside GhSWEET14a/b. CRISPR-Cas9 gene editing supports the function of GhSWEETs in cotton bacterial blight and the promise of disrupting the TALE-binding site in S genes for disease management. Collectively, our findings elucidate the rapid evolution of TALEs in Xanthomonas field isolates and highlight the virulence mechanism wherein TALEs induce multiple S genes to promote pathogenicity. Newly evolved Xanthomonas citri pv. malvacearum isolates triggers recent bacterial blight outbreaks in cotton. Here, the authors show that a recently evolved TALE, Tal7b, activates host susceptibility genes GhSWEET14a and GhSWEET14b rather than GhSWEET10 to confer pathogenicity in these new isolates.

|
Scooped by
dromius
January 19, 10:24 AM
|
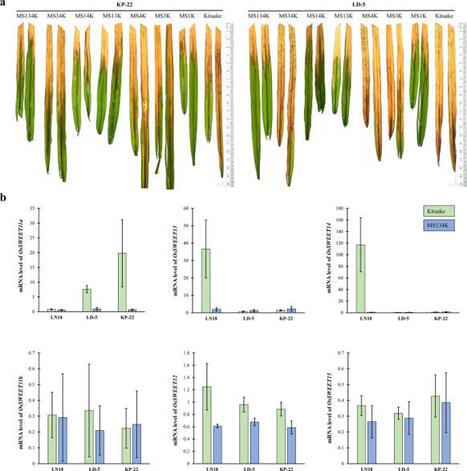
Syed Mashab Ali Shah et al. 2025
Bacterial blight (BB) of rice caused by Xanthomonas oryzae pv. oryzae (Xoo), is an important disease in rice-growing countries, including Pakistan, where it was first reported in the mid-1970s. Transcription activator-like effectors (TALEs) play vital roles in many plant diseases caused by Xanthomonas spp.; however, Pakistani Xoo TALome diversity and their contribution to pathogenicity is largely unknown. In this study, 101 Xoo strains were screened using specific PCR primers. The genomic DNA from these strains underwent BamHI digestion and hybridized with the internal SphI fragment of PthXo1. Southern blot analysis revealed 16 to 20 putative tale fragments among the tested strains. These strains were further classified into 11 genotypes based on the number and size of the hybridizing bands. Genotypes 1, 2, 3, and 4 represented 24, 2, 51, and 17 strains, respectively. Pathogenicity assays on near-isogenic lines (NILs) containing different resistance (R) genes exhibited that CBB23 was incompatible with all tested Pakistani-Xoo genotypes, whereas IRBB5 and IRBB4 showed resistance against specific genotypes. In contrast, paddy trails on NILs containing single, double, and triple mutants of OsSWEET11a, OsSWEET13, and OsSWEET14 in the effector binding elements (EBEs) of cv. Kitaake revealed that KP-22 and LD-5 harbor novel virulent TAL effector/s. Interestingly, the expression analysis of six clade-III OsSWEET genes suggests that novel TALE/s targeting unidentified susceptibility gene/s. Altogether, this study highlights gene-for-gene relationships between tested rice lines and Pakistani-Xoo strains. This is the first report providing the diversity of TALEs and their relationship to R and S (susceptibility) genes. Further identification of novel virulent TALE/s and their cognate target/s is warranted to precisely elucidate their role in BB.

|
Scooped by
dromius
October 27, 2024 7:37 AM
|
Gaudin et al. 2024
Common bacterial blight of bean (CBB) is a devastating seed-transmitted disease caused by Xanthomonas phaseoli pv. phaseoli and Xanthomonas citri pv. fuscans on common bean (Phaseolus vulgaris L.). The genes responsible for CBB resistance are largely unknown. Moreover, the lack of a reproducible and universal transformation protocol limits the study of genetic traits in common bean. We produced X. phaseoli pv. phaseoli strains expressing artificially-designed Transcription-Activator Like Effectors (dTALEs) to target 14 candidate genes for resistance to CBB based on previous transcriptomic data. In planta assays in a susceptible common bean genotype showed that induction of PvOFP7, PvAP2‐ERF71 or PvExpansinA17 expression by dTALEs resulted in CBB symptom reduction. After PvOFP7 induction, in planta bacterial growth was reduced at early colonisation stages and RNA-Seq analysis revealed up-regulation of cell wall formation and primary metabolism, together with major down-regulation of Heat Shock Proteins. Our results demonstrate that PvOFP7 contributes to CBB resistance, and underline the usefulness of dTALEs for functional validation of genes whose induction impacts Xanthomonas-plant interaction.

|
Scooped by
dromius
May 31, 2024 4:33 AM
|
Liu et al. 2024
N6-methyladenosine (6mA) plays a pivotal role in diverse biological processes, including cancer, bacterial toxin secretion, and bacterial drug resistance. However, to date there has not been a selective, sensitive, and simple method for quantitative detection of 6mA at single base resolution. Herein, we present a series piezoelectric quartz crystal (SPQC) sensor based on the specific recognition of transcription-activator-like effectors (TALEs) for locus-specific detection of 6mA. Detection sensitivity is enhanced through the use of a hybridization chain reaction (HCR) in conjunction with silver staining. The limit of detection (LOD) of the sensor was 0.63 pM and can distinguish single base mismatches. We demonstrate the applicability of the sensor platform by quantitating 6mA DNA at a specific site in biological matrix. The SPQC sensor presented herein offers a promising platform for in-depth study of cancer, bacterial toxin secretion, and bacterial drug resistance.

|
Scooped by
dromius
April 30, 2024 3:20 AM
|
Zhang et al. 2024 Background
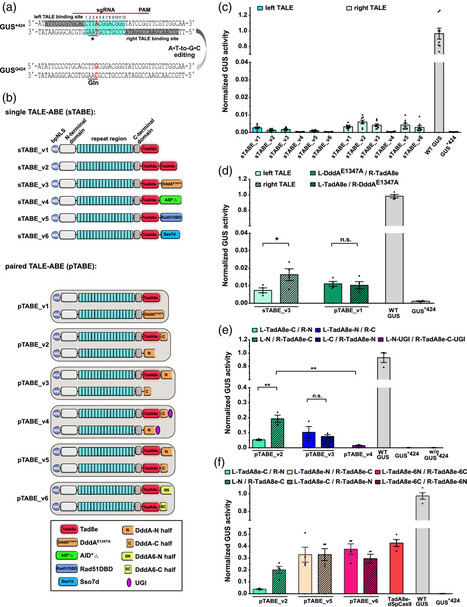
TALE-derived DddA-based cytosine base editors (TALE-DdCBEs) can perform efficient base editing of mitochondria and chloroplast genomes. They use transcription activator-like effector (TALE) arrays as programmable DNA-binding domains and a split version of the double-strand DNA cytidine deaminase (DddA) to catalyze C•G-to-T•A editing. This technology has not been optimized for use in plant cells. Results
To systematically investigate TALE-DdCBE architectures and editing rules, we established a β-glucuronidase reporter for transient assays in Nicotiana benthamiana. We show that TALE-DdCBEs function with distinct spacer lengths between the DNA-binding sites of their two TALE parts. Compared to canonical DddA, TALE-DdCBEs containing evolved DddA variants (DddA6 or DddA11) showed a significant improvement in editing efficiency in Nicotiana benthamiana and rice. Moreover, TALE-DdCBEs containing DddA11 have broader sequence compatibility for non-TC target editing. We have successfully regenerated rice with C•G-to-T•A conversions in their chloroplast genome, as well as N. benthamiana with C•G-to-T•A editing in the nuclear genome using TALE-DdCBE. We also found that the spontaneous assembly of split DddA halves can cause undesired editing by TALE-DdCBEs in plants.
Conclusions
Altogether, our results refined the targeting scope of TALE-DdCBEs and successfully applied them to target the chloroplast and nuclear genomes. Our study expands the base editing toolbox in plants and further defines parameters to optimize TALE-DdCBEs for high-fidelity crop improvement.

|
Scooped by
dromius
January 17, 2024 1:51 AM
|
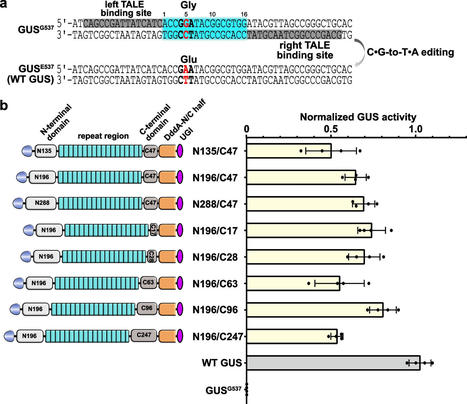
Zhang & Boch 2023 Base editors enable precise nucleotide changes at targeted genomic loci without requiring double-stranded DNA breaks or repair templates. TALE-adenine base editors (TALE-ABEs) are genome editing tools, composed of a DNA-binding domain from transcription activator-like effectors (TALEs), an engineered adenosine deaminase (TadA8e), and a cytosine deaminase domain (DddA), that allow A•T-to-G•C editing in human mitochondrial DNA. However, the editing ability of TALE-ABEs in plants apart from chloroplast DNA has not been described, so far, and the functional role how DddA enhances TadA8e is still unclear. We tested a series of TALE-ABEs with different deaminase fusion architectures in Nicotiana benthamiana and rice. The results indicate that the double-stranded DNA-specific cytosine deaminase DddA can boost the activities of single-stranded DNA-specific deaminases (TadA8e or APOBEC3A) on double-stranded DNA. We analysed A•T-to-G•C editing efficiencies in a β-glucuronidase reporter system and showed precise adenine editing in genomic regions with high product purity in rice protoplasts. Furthermore, we have successfully regenerated rice plants with A•T-to-G•C mutations in the chloroplast genome using TALE-ABE. Consequently, TALE-adenine base editors provide alternatives for crop improvement and gene therapy by editing nuclear or organellar genomes.

|
Scooped by
dromius
November 7, 2023 2:21 PM
|
You and Jiang, 2023
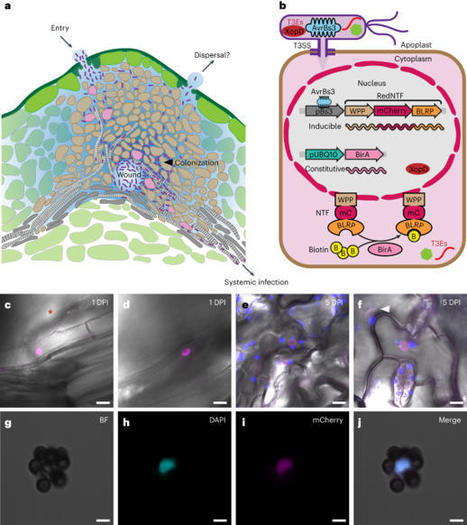
Type-III effector proteins are major virulence determinants that most gram-negative bacteria inject into host cells to manipulate cellular processes for infection. Because effector-targeted cells are embedded and underrepresented in infected plant tissues, it is technically challenging to isolate them for focused studies of effector-induced cellular changes.
This protocol describes a novel technique, effector-inducible isolation of nuclei tagged in specific cell types (eINTACT), for isolating biotin-labeled nuclei from Arabidopsis plant cells that have received Xanthomonas bacterial effectors by using streptavidin-coated magnetic beads. This protocol is an extension of the existing Nature Protocols Protocol of the INTACT method for the affinity-based purification of nuclei of specific cell types in the context of developmental biology. In a phytopathology scenario, our protocol addresses how to obtain eINTACT transgenic lines and compatible bacterial mutants, verify the eINTACT system and purify nuclei of bacterial effector-recipient cells from infected tissues.
Differential analyses of purified nuclei from plants infected by bacteria expressing the effector of interest and those from plants infected by effector-deletion bacterial mutants will reveal the effector-dependent nuclear changes in targeted host cells. Provided that the eINTACT system is available, the infection experiment takes 5 d, and the procedures, from collecting bacteria-infected leaves to obtaining nuclei of effector-targeted cells, can be completed in 4 h.
eINTACT is a unique method for isolating high-quality nuclei from bacterial effector-targeted host cells in native infection contexts. This method is adaptable to study the functions of type-III effectors from numerous gram-negative bacteria in host plants that are amenable to transformation.
|




 Your new post is loading...
Your new post is loading...