Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
February 14, 2013 1:25 PM
|
(via Tom Schreiber. Thanks!) Mashimo et al, 2013 TAL Effector Nucleases (TALENs) are versatile tools for targeted gene editing in various species. However, their efficiency is still insufficient, especially in mammalian embryos. Here, we showed that combined expression of Exonuclease 1 (Exo1) with engineered site-specific TALENs provided highly efficient disruption of the endogenous gene in rat fibroblast cells. A similar increased efficiency of up to ~30% with Exo1 was also observed in fertilized rat eggs, and in the production of knockout rats for the albino (Tyr) gene. These findings demonstrate TALENs with Exo1 is an easy and efficient method of generating gene knockouts using zygotes, which increases the range of gene targeting technologies available to various species.

|
Scooped by
dromius
February 7, 2013 9:47 AM
|
via Tom Schreiber, thanks. Sakuma et al, 2013 Transcription activator–like effector nucleases (TALENs) have recently arisen as effective tools for targeted genome engineering. Here, we report streamlined methods for the construction and evaluation of TALENs based on the ‘Golden Gate TALEN and TAL Effector Kit’ (Addgene). We diminished array vector requirements and increased assembly rates using six-module concatemerization. We altered the architecture of the native TALEN protein to increase nuclease activity and replaced the final destination vector with a mammalian expression/in vitro transcription vector bearing both CMV and T7 promoters. Using our methods, the whole process, from initiating construction to completing evaluation directly in mammalian cells, requires only 1 week. Furthermore, TALENs constructed in this manner may be directly applied to transfection of cultured cells or mRNA synthesis for use in animals and embryos. In this article, we show genomic modification of HEK293T cells, human induced pluripotent stem cells, Drosophila melanogaster, Danio rerio and Xenopus laevis, using custom-made TALENs constructed and evaluated with our protocol. Our methods are more time efficient compared with conventional yeast-based evaluation methods and provide a more accessible and effective protocol for the application of TALENs in various model organisms.

|
Scooped by
dromius
February 4, 2013 3:06 AM
|
http://www.talendesign.org/mojohand_main.php We present the web-based program Mojo Hand for designing TAL and TALEN constructs for genome editing applications (www.talendesign.org). We describe the algorithm and its implementation. The features of Mojo Hand include (1) automatic download of genomic data from the National Center for Biotechnology Information, (2) analysis of any DNA sequence to reveal pairs of binding sites based on a user-defined template, (3) selection of restriction enzyme recognition sites in the spacer between the TAL monomer binding sites including options for the selection of restriction enzyme suppliers, and (4) output files designed for subsequent TALEN construction using the Golden Gate assembly method.

|
Scooped by
dromius
February 4, 2013 3:00 AM
|
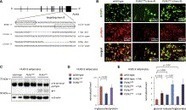
Background: Human mitochondrial complex I is composed of 44 subunits with an elaborate assembly mechanism. Results: TALENs were used to knock out complex I subunit NDUFA9 in HEK293T cells, revealing a novel membrane arm subcomplex. Conclusion: NDUFA9 is critical for stabilizing the junction between different arms of complex I. Significance: TALEN-mediated knock-out represents an accessible approach to address protein function in human cells.

|
Scooped by
dromius
January 12, 2013 6:48 PM
|
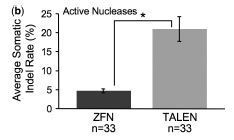
via Tom Schreiber, thanks Chen et al, 2013 Zinc-finger nucleases (ZFNs) and TAL effector nucleases (TALENs) have been shown to induce targeted mutations, but they have not been extensively tested in any animal model. Here, we describe a large-scale comparison of ZFN and TALEN mutagenicity in zebrafish. Using deep sequencing, we found that TALENs are significantly more likely to be mutagenic and induce an average of 10-fold more mutations than ZFNs. We observed a strong correlation between somatic and germ-line mutagenicity, and identified germ line mutations using ZFNs whose somatic mutations rates are well below the commonly used threshold of 1%. Guidelines that have previously been proposed to predict optimal ZFN and TALEN target sites did not predict mutagenicity in vivo. However, we observed a significant negative correlation between TALEN mutagenicity and the number of CpG repeats in TALEN target sites, suggesting that target site methylation may explain the poor mutagenicity of some TALENs in vivo. The higher mutation rates and ability to target essentially any sequence make TALENs the superior technology for targeted mutagenesis in zebrafish, and likely other animal models.

|
Scooped by
dromius
January 7, 2013 12:38 PM
|
Four rice genes and eight Brachypodium genes were targeted to generate knockout mutations. A protoplast transient assay was developed in both rice and Brachypodium. TALEN-encoding constructs were introduced into protoplasts using polyethylene glycol (PEG). Seven out of eleven TALENs showed mutagenesis frequencies ranging from
4% to 14%. The TALENs targeting the 4 rice and 8 Brachypodium genes were transformed into plants. The constructs were introduced into embryonic cells of rice or Brachypodium using Agrobacterium tumefaciens. Two rice TALENs separated by 1322 bp in the OsBADH2
gene, were co-introduced into rice protoplasts. These two TALEN pairs were next co-delivered into rice calli by particle bombardment. Ninety-eight hygromycin-resistant calli were screened after 6 weeks of selection, and three types of modifications were identified — simple modifications (small deletions or insertions) at either target site, large deletions of the sequence between the
two target sites and inversion of the sequence between the two target sites. These results support the idea that TALEN-induced genomic deletions were mediated via NHEJ. Targeted deletions would make possible the selective removal of gene clusters and enables scientists to delete intergenic regions, introns, regulatory elements and noncoding RNAs.
To our knowledge, this is the first study demonstrating highly efficient targeted
knockouts in multiple genes in the model monocot species Brachypodium. We anticipate TALEN technology will make targeted gene modification a routine practice not just for these model monocots but also for economically important crops, such as maize and wheat.

|
Suggested by
Qizhang Long
January 5, 2013 7:13 AM
|

|
Scooped by
dromius
December 27, 2012 9:37 PM
|

|
Scooped by
dromius
December 23, 2012 10:19 PM
|

|
Suggested by
Pawel Bialk
December 18, 2012 4:35 PM
|

|
Scooped by
dromius
December 12, 2012 3:26 AM
|
The Two Blades Foundation (2Blades) announced the completion of a non-exclusive license agreement with DuPont Pioneer for access to 2Blades' TAL Code technology for commercial uses in certain crop plants.

|
Suggested by
Qizhang Long
December 7, 2012 9:50 AM
|
EENdb: a database and knowledge base of ZFNs and TALENs for endonuclease engineering

|
Suggested by
asc444
November 28, 2012 7:11 PM
|
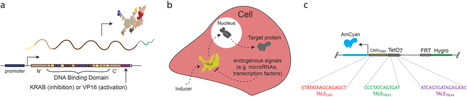
Thanks, Anita! Li et al. (2012) The ability to conditionally rewire pathways in human cells holds great therapeutic potential. Transcription activator-like effectors (TALEs) are a class of naturally occurring specific DNA binding proteins that can be used to introduce targeted genome modifications or control gene expression. Here we present TALE hybrids engineered to respond to endogenous signals and capable of controlling transgenes by applying a predetermined and tunable action at the single-cell level. Specifically, we first demonstrate that combinations of TALEs can be used to modulate the expression of stably integrated genes in kidney cells. We then introduce a general purpose two-hybrid approach that can be customized to regulate the function of any TALE either using effector molecules or a heterodimerization reaction. Finally, we demonstrate the successful interface of TALEs to specific endogenous signals, namely hypoxia signaling and microRNAs, essentially closing the loop between cellular information and chromosomal transgene expression.
|

|
Scooped by
dromius
February 13, 2013 11:24 AM
|
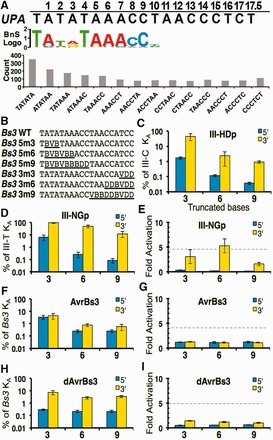
(via Tom Schreiber, thanks!) Meckler et al, 2013 Transcription activator-like effectors (TALEs) have revolutionized the field of genome engineering. We present here a systematic assessment of TALE DNA recognition, using quantitative electrophoretic mobility shift assays and reporter gene activation assays. Within TALE proteins, tandem 34-amino acid repeats recognize one base pair each and direct sequence-specific DNA binding through repeat variable di-residues (RVDs). We found that RVD choice can affect affinity by four orders of magnitude, with the relative RVD contribution in the order NG > HD ∼ NN ≫ NI > NK. The NN repeat preferred the base G over A, whereas the NK repeat bound G with 103-fold lower affinity. We compared AvrBs3, a naturally occurring TALE that recognizes its target using some atypical RVD-base combinations, with a designed TALE that precisely matches ‘standard’ RVDs with the target bases. This comparison revealed unexpected differences in sensitivity to substitutions of the invariant 5′-T. Another surprising observation was that base mismatches at the 5′ end of the target site had more disruptive effects on affinity than those at the 3′ end, particularly in designed TALEs. These results provide evidence that TALE–DNA recognition exhibits a hitherto un-described polarity effect, in which the N-terminal repeats contribute more to affinity than C-terminal ones.

|
Scooped by
dromius
February 4, 2013 11:56 AM
|
via Tom Schreiber (thanks!) Perez-Pinera et al, 2013 Mammalian genes are regulated by the cooperative and synergistic actions of many transcription factors. In this study we recapitulate this complex regulation in human cells by targeting endogenous gene promoters, including regions of closed chromatin upstream of silenced genes, with combinations of engineered transcription activator–like effectors (TALEs). These combinations of TALE transcription factors induced substantial gene activation and allowed tuning of gene expression levels that will broadly enable synthetic biology, gene therapy and biotechnology.

|
Scooped by
dromius
February 4, 2013 3:03 AM
|
We streamlined a Golden Gate-based method for custom TALE assembly. First, by providing ready-made, quality-controlled monomers, we eliminated the procedures for error-prone and time-consuming set-up. Second, we optimized the protocol toward a fast, two-day assembly of custom TALEs, based on four thermocycling reactions. Third, we increased the versatility for diverse downstream applications by providing series of vector sets to generate both TALENs (TALE nucleases) and TALE-TFs (TALE-transcription factors) under the control of different promoters. Finally, we validated our system by assembling a number of TALENs and TALE-TFs with DNA sequencing confirmation. We further demonstrated that an assembled TALE-TF was able to transactivate a luciferase reporter gene and a TALEN pair was able to cut its target.

|
Scooped by
dromius
January 16, 2013 3:24 AM
|
Kim et al, 2013 Xo2276 is a putative transcription activator-like effector (TALE) in Xanthomonas oryzae pv. oryzae (Xoo). Xo2276 was expressed with a TAP-tag at the C-terminus in Xoo cells to enable quantitative analysis of protein expression and secretion. Nearly all TAP-tagged Xo2276 existed in an insoluble form; addition of rice leaf extracts from a Xoosusceptible rice cultivar, Milyang23, significantly stimulated secretion of TAP-tagged Xo2276 into the medium. In a T3SS-defective Xoo mutant strain, secretion of TAPtagged Xo2276 was blocked. Xo2276 is a Xoo ortholog of Xanthomonas campestris pv. vesicatoria (Xcv) AvrBs3 and contains a conserved DNA-binding domain (DBD), which includes 19.5 tandem repeats of 34 amino acids. Xo2276- DBD was expressed in E. coli and purified. Direct in vitro recognition of Xo2276-DBD on a putative target DNA sequence was confirmed using an electrophoretic mobility shift assay. This is the first study measuring the homologous expression and secretion of Xo2276 in vitro using rice leaf extract and its direct in vitro binding to the specific target DNA sequence.

|
Scooped by
dromius
January 11, 2013 3:49 PM
|
Bochtler 2012 Phytopathogen transcription activator-like effectors (TALEs) bind DNA in a sequence specific manner in order to manipulate host transcription. TALE specificity correlates with repeat variable diresidues in otherwise highly stereotypical 34–35mer repeats. Recently, the crystal structures of two TALE DNA-binding domains have illustrated the molecular basis of the TALE cipher. The structures show that the TALE repeats form a right-handed superhelix that is wound around largely undistorted B-DNA to match its helical parameters. Surprisingly, repeat variable residue 1 is not in contact with the bases. Instead, it is involved in hydrogen bonding interactions that stabilize the overall structure of the protein. Repeat variable residue 2 contacts the top strand base and forms sequence-specific hydrogen bonds and/or van der Waals contacts. Very unexpectedly, bottom strand bases are exposed to solvent and do not make any direct contacts with the protein. This review contains a summary of TALE biology and applications and a detailed description of the recent breakthroughs that have provided insights into the molecular basis of the TALE code.

|
Scooped by
dromius
January 5, 2013 7:25 AM
|
8. A new tool helps researchers manipulate our genes.
In 2012 researchers created a remarkable protein that they can use to change a cell's genes on the fly. Known as TALENs, which stands for "transcription activator-like effector nucleases," gave researchers the ability to alter or inactivate specific genes in zebrafish, toads, livestock and other animals — even cells from patients with disease. These TALENs can be customized to seek out and edit any DNA sequence, so it's very specific. It can be used to chop out a bad gene and replace it for a treatment, or can be used in research to understand what specific DNA stretches do in healthy and diseased individuals.

|
Scooped by
dromius
December 27, 2012 10:30 PM
|

|
Scooped by
dromius
December 23, 2012 10:25 PM
|

|
Scooped by
dromius
December 21, 2012 12:13 AM
|
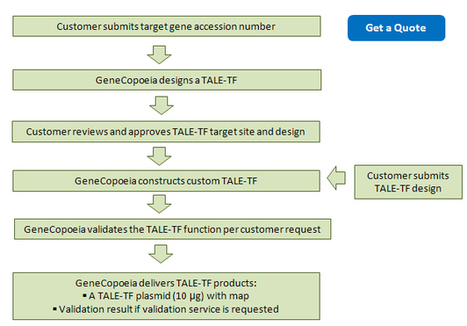
GeneCopoeia offers custom services for designing, creating and validating TALENs, TALE-TFs and other TAL effectors-based targeted genomic modification tools.

|
Rescooped by
dromius
from Plants and Microbes
December 14, 2012 12:53 PM
|
Biologists have turned plant pest proteins into tools for studying and reshaping genomes of many species. Some of biology's best technologies come from unexpected places. The green fluorescent protein that lit up biology with its ability to track proteins and gene expression in cells was borrowed from a jellyfish. A heat-stable enzyme from a bacterium often found in hot springs made the polymerase chain reaction method practical, facilitating the easy copying of DNA fragments needed for a myriad of applications, including the DNA fingerprinting used so widely to identify people. Now, thanks in part to inspiration that struck during a lunchtime discussion, proteins from a feared plant pest are poised to make genome engineering, the large-scale, directed manipulation of genes, routine for researchers studying a variety of organisms, including yeast and humans.
Via Kamoun Lab @ TSL

|
Scooped by
dromius
December 7, 2012 9:53 AM
|
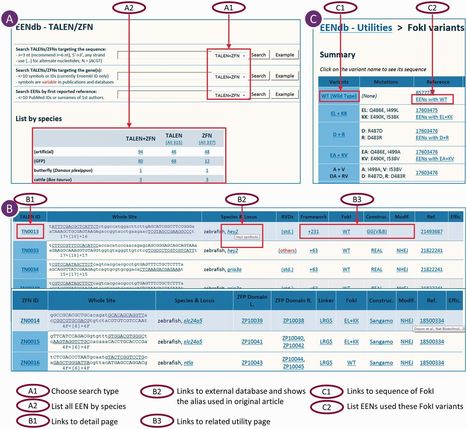
We report here the construction of engineered endonuclease database (EENdb) (http://eendb.zfgenetics.org/), a searchable database and knowledge base for customizable engineered endonucleases (EENs), including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). EENs are artificial nucleases designed to target and cleave specific DNA sequences. EENs have been shown to be a very useful genetic tool for targeted genome modification and have shown great potentials in the applications in basic research, clinical therapies and agricultural utilities, and they are specifically essential for reverse genetics research in species where no other gene targeting techniques are available. EENdb contains over 700 records of all the reported ZFNs and TALENs and related information, such as their target sequences, the peptide components [zinc finger protein-/transcription activator-like effector (TALE)-binding domains, FokI variants and linker peptide/framework], the efficiency and specificity of their activities. The database also lists EEN engineering tools and resources as well as information about forms and types of EENs, EEN screening and construction methods, detection methods for targeting efficiency and many other utilities. The aim of EENdb is to represent a central hub for EEN information and an integrated solution for EEN engineering. These studies may help to extract in-depth properties and common rules regarding ZFN or TALEN efficiency through comparison of the known ZFNs or TALENs.

|
Scooped by
dromius
November 28, 2012 7:14 PM
|
RT @JBiolChem: Paper of the week: Overcoming TALE DNA Binding Domain's Inability to Bind Methylcytosine http://t.co/L8cs2yHV...
|



 Your new post is loading...
Your new post is loading...