Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
October 12, 2022 6:18 PM
|
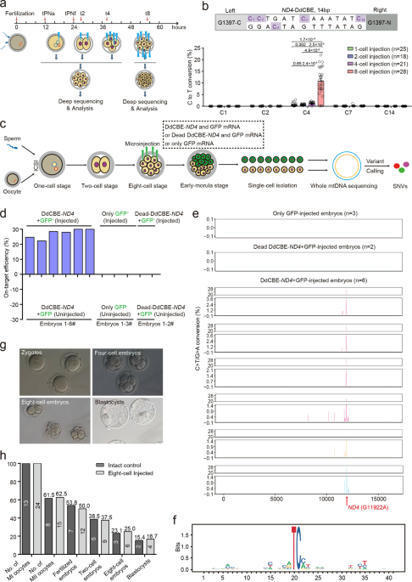
(via T. Lahaye, thx) Lee et al. 2022 We present two methods for enhancing the efficiency of mitochondrial DNA (mtDNA) editing in mice with DddA-derived cytosine base editors (DdCBEs). First, we fused DdCBEs to a nuclear export signal (DdCBE-NES) to avoid off-target C-to-T conversions in the nuclear genome and improve editing efficiency in mtDNA. Second, mtDNA-targeted TALENs (mitoTALENs) are co-injected into mouse embryos to cleave unedited mtDNA. We generated a mouse model with the m.G12918A mutation in the MT-ND5 gene, associated with mitochondrial genetic disorders in humans. The mutant mice show hunched appearances, damaged mitochondria in kidney and brown adipose tissues, and hippocampal atrophy, resulting in premature death.

|
Scooped by
dromius
September 3, 2022 11:06 AM
|
(via T. Lahaye, thx)
Yoshihisa et al, 2022
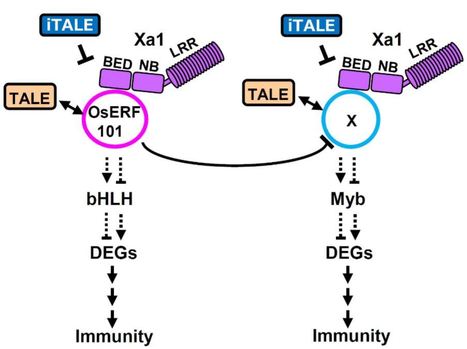
Plant nucleotide-binding leucine-rich repeat receptors (NLRs) initiate immune responses by recognizing pathogen effectors. The rice gene Xa1 encodes an NLR with an N-terminal BED domain, and recognizes transcription activator-like (TAL) effectors of Xanthomonas oryzae pv. oryzae (Xoo). Our goal is to elucidate the molecular mechanisms controlling the induction of immunity by Xa1.
We used yeast two-hybrid assays to screen for host factors that interact with Xa1 and identified the AP2/ERF-type transcription factor OsERF101/OsRAP2.6. Molecular complementation assays were used to confirm the interactions among Xa1, OsERF101, and two TAL effectors. We created OsERF101-overexpressing and knockout mutant lines in rice and identified genes differentially regulated in these lines, many of which are predicted to be involved in regulation of response to stimulus.
Xa1 interacts in the nucleus with the TAL effectors and OsERF101 via the BED domain. Unexpectedly, both the overexpression and knockout lines of OsERF101 displayed Xa1-dependent, enhanced resistance to an incompatible Xoo strain. Different sets of genes were up- or down-regulated in the overexpression and knockout lines.
Our results indicate that OsERF101 regulates the recognition of TAL effectors by Xa1, and functions as a positive regulator of Xa1-mediated immunity. Further, an additional Xa1-mediated immune pathway is negatively regulated by OsERF101.

|
Scooped by
dromius
August 5, 2022 1:24 PM
|
(via T. Schreiber, thx) Roeschlin et al, 2019 Transcription activator-like effectors (TALEs) are important effectors of Xanthomonas spp. that manipulate the transcriptome of the host plant, conferring susceptibility or resistance to bacterial infection. Xanthomonas citri ssp. citri variant AT (X. citri AT) triggers a host-specific hypersensitive response (HR) that suppresses citrus canker development. However, the bacterial effector that elicits this process is unknown. In this study, we show that a 7.5-repeat TALE is responsible for triggering the HR. PthA4AT was identified within the pthA repertoire of X. citri AT followed by assay of the effects on different hosts. The mode of action of PthA4AT was characterized using protein-binding microarrays and testing the effects of deletion of the nuclear localization signals and activation domain on plant responses. PthA4AT is able to bind DNA and activate transcription in an effector binding element-dependent manner. Moreover, HR requires PthA4AT nuclear localization, suggesting the activation of executor resistance (R) genes in host and non-host plants. This is the first case where a TALE of unusually short length performs a biological function by means of its repeat domain, indicating that the action of these effectors to reprogramme the host transcriptome following nuclear localization is not limited to ‘classical’ TALEs.

|
Scooped by
dromius
July 13, 2022 6:35 AM
|
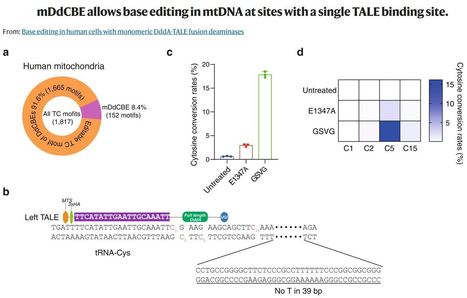
(via T. Schreiber, thx) Young Geun Mok et al. 2022 Inter-bacterial toxin DddA-derived cytosine base editors (DdCBEs) enable targeted C-to-T conversions in nuclear and organellar DNA. DddAtox, the deaminase catalytic domain derived from Burkholderia cenocepacia, is split into two inactive halves to avoid its cytotoxicity in eukaryotic cells, when fused to transcription activator-like effector (TALE) DNA-binding proteins to make DdCBEs. As a result, DdCBEs function as pairs, which hampers gene delivery via viral vectors with a small cargo size. Here, we present non-toxic, full-length DddAtox variants to make monomeric DdCBEs (mDdCBEs), enabling mitochondrial DNA editing with high efficiencies of up to 50%, when transiently expressed in human cells. We demonstrate that mDdCBEs expressed via AAV in cultured human cells can achieve nearly homoplasmic C-to-T editing in mitochondrial DNA. Interestingly, mDdCBEs often produce mutation patterns different from those obtained with conventional dimeric DdCBEs. Furthermore, mDdCBEs allow base editing at sites for which only one TALE protein can be designed. We also show that transfection of mDdCBE-encoding mRNA, rather than plasmid, can reduce off-target editing in human mitochondrial DNA.

|
Scooped by
dromius
July 1, 2022 4:13 AM
|
Danila et al. 2022
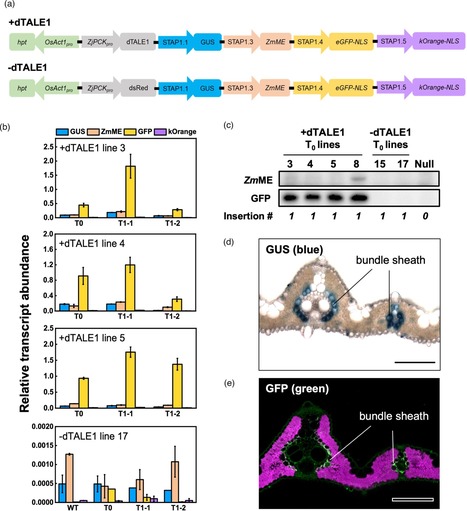
In biological discovery and engineering research, there is a need to spatially and/or temporally regulate transgene expression. However, the limited availability of promoter sequences that are uniquely active in specific tissue-types and/or at specific times often precludes co-expression of multiple transgenes in precisely controlled developmental contexts. Here, we developed a system for use in rice that comprises synthetic designer transcription activator-like effectors (dTALEs) and cognate synthetic TALE-activated promoters (STAPs). The system allows multiple transgenes to be expressed from different STAPs, with the spatial and temporal context determined by a single promoter that drives expression of the dTALE. We show that two different systems—dTALE1-STAP1 and dTALE2-STAP2—can activate STAP-driven reporter gene expression in stable transgenic rice lines, with transgene transcript levels dependent on both dTALE and STAP sequence identities. The relative strength of individual STAP sequences is consistent between dTALE1 and dTALE2 systems but differs between cell-types, requiring empirical evaluation in each case. dTALE expression leads to off-target activation of endogenous genes but the number of genes affected is substantially less than the number impacted by the somaclonal variation that occurs during the regeneration of transformed plants. With the potential to design fully orthogonal dTALEs for any genome of interest, the dTALE-STAP system thus provides a powerful approach to fine-tune the expression of multiple transgenes, and to simultaneously introduce different synthetic circuits into distinct developmental contexts.

|
Scooped by
dromius
April 26, 2022 2:48 AM
|
Mishra et al. 2022
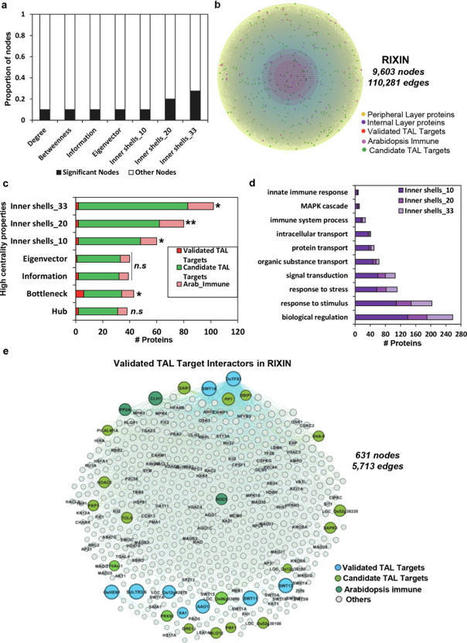
Network science identifies key players in diverse biological systems including host-pathogen interactions. We demonstrated a scale-free network property for a comprehensive rice protein-protein interactome (RicePPInets) that exhibits nodes with increased centrality indices. While weighted k-shell decomposition was shown efficacious to predict pathogen effector targets in Arabidopsis, we improved its computational code for a broader implementation on large-scale networks including RicePPInets. We determined that nodes residing within the internal layers of RicePPInets are poised to be the most influential, central, and effective information spreaders. To identify central players and modules through network topology analyses, we integrated RicePPInets and co-expression networks representing susceptible and resistant responses to strains of the bacterial pathogens Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola (Xoc) and generated a RIce-Xanthomonas INteractome (RIXIN). This revealed that previously identified candidate targets of pathogen transcription activator-like (TAL) effectors are enriched in nodes with enhanced connectivity, bottlenecks, and information spreaders that are located in the inner layers of the network, and these nodes are involved in several important biological processes. Overall, our integrative multi-omics network-based platform provides a potentially useful approach to prioritizing candidate pathogen effector targets for functional validation, suggesting that this computational framework can be broadly translatable to other complex pathosystems.

|
Scooped by
dromius
March 18, 2022 1:34 AM
|
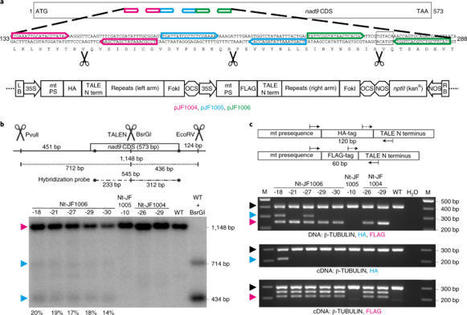
Forner et al. 2022 The development of technologies for the genetic manipulation of mitochondrial genomes remains a major challenge. Here we report a method for the targeted introduction of mutations into plant mitochondrial DNA (mtDNA) that we refer to as transcription activator-like effector nuclease (TALEN) gene-drive mutagenesis (GDM), or TALEN-GDM. The method combines TALEN-induced site-specific cleavage of the mtDNA with selection for mutations that confer resistance to the TALEN cut. Applying TALEN-GDM to the tobacco mitochondrial nad9 gene, we isolated a large set of mutants carrying single amino acid substitutions in the Nad9 protein. The mutants could be purified to homochondriomy and stably inherited their edited mtDNA in the expected maternal fashion. TALEN-GDM induces both transitions and transversions, and can access most nucleotide positions within the TALEN binding site. Our work provides an efficient method for targeted mitochondrial genome editing that produces genetically stable, homochondriomic and fertile plants with specific point mutations in their mtDNA. A method for targeted mutagenesis of mitochondrial genomes is presented. It combines site-specific DNA cleavage with selection for mutations that confer cleavage resistance, and produces genetically stable plants with edited mitochondrial genomes.

|
Scooped by
dromius
March 6, 2022 5:07 PM
|
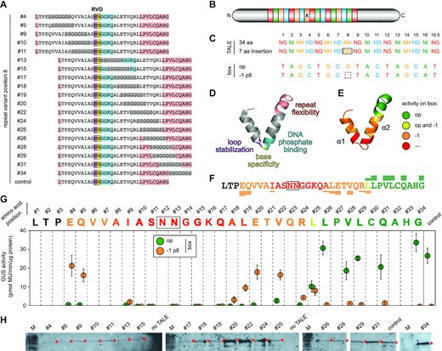
Becker et al, 2022 Transcription activator-like effectors (TALEs) are bacterial proteins with a programmable DNA-binding domain, which turned them into exceptional tools for biotechnology. TALEs contain a central array of consecutive 34 amino acid long repeats to bind DNA in a simple one-repeat-to-one-nucleotide manner. However, a few naturally occurring aberrant repeat variants break this strict binding mechanism, allowing for the recognition of an additional sequence with a −1 nucleotide frameshift. The limits and implications of this extended TALE binding mode are largely unexplored. Here, we analyse the complete diversity of natural and artificially engineered aberrant repeats for their impact on the DNA binding of TALEs. Surprisingly, TALEs with several aberrant repeats can loop out multiple repeats simultaneously without losing DNA-binding capacity. We also characterized members of the only natural TALE class harbouring two aberrant repeats and confirmed that their target is the major virulence factor OsSWEET13 from rice. In an aberrant TALE repeat, the position and nature of the amino acid sequence strongly influence its function. We explored the tolerance of TALE repeats towards alterations further and demonstrate that inserts as large as GFP can be tolerated without disrupting DNA binding. This illustrates the extraordinary DNA-binding capacity of TALEs and opens new uses in biotechnology.

|
Scooped by
dromius
February 6, 2022 8:28 AM
|
Dinh et al, 2021 - Preprint
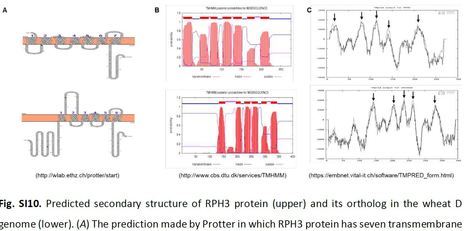
Host resistance is considered the most effective means to control plant diseases; however, individually deployed resistance genes are often rapidly overcome by pathogen adaptation. Combining multiple effective resistance genes is the optimal approach to durable resistance, but the lack of functional markers for resistance genes has hampered implementation. Leaf rust, caused by Puccinia hordei, is an economically significant disease of barley, but only a few major Resistance genes to P. hordei (Rph) have been cloned. In this study, gene Rph3 was isolated by positional cloning and confirmed by mutational analysis and transgenic complementation. The Rph3 gene, which originated from wild barley and was first introgressed into cultivated Egyptian germplasm, encodes a unique transmembrane resistance protein that differs from all known plant disease resistance proteins at the amino acid sequence level. Genetic profiles of diverse accessions indicated limited genetic diversity in Rph3 in domesticated germplasm, and higher diversity in wild barley from the Eastern Mediterranean region. Expression profiling using P. hordei isolates with contrasting pathogenicity for the Rph3 host locus showed that the Rph3 gene was expressed only in interactions with Rph3-avirulent isolates, a phenomenon also observed for transcription activator-like effector-dependent genes known as executors conferring resistance to Xanthomonas spp. Like the known transmembrane executors such as Bs3 and Xa7 heterologous expression of Rph3 in N. benthamiana induced a cell death response. Given that Rph3 shares several features with executor genes, it seems likely that P. hordei contains effectors similar to the transcription activator-like effectors that target host executor genes. The isolation of Rph3 highlights convergent evolutionary processes in diverse plant-pathogen interaction systems, where similar defence mechanisms evolved independently in monocots and dicots and provide evidence for executor genes in the Triticeae tribe.

|
Scooped by
dromius
February 3, 2022 6:31 AM
|
(via T. Lahaye, thx) Cowles et al, 2022 - Preprint
Salmonella enterica is ubiquitous in the plant environment, persisting in the face of UV stress, plant defense responses, desiccation, and nutrient limitation. These fluctuating conditions of the leaf surface result in S. enterica population decline. Biomultipliers, such as the phytopathogenic bacterium Xanthomonas hortorum pv. gardneri, alter the phyllosphere to the benefit of S. enterica. Specific X. gardneri-dependent changes to this niche that promote S. enterica persistence remain unclear, and this work focuses on identifying factors that lead to increased S. enterica survival on leaves. Here, we show that the X. gardneri transcription activator-like effector AvrHah1 is both necessary and sufficient for increased survival of S. enterica on tomato leaves. An X. gardneri avrHah1 mutant fails to influence S. enterica survival while addition of avrHah1 to X. vesicatoria provides a gain of function. Our results indicate that although X. gardneri stimulates a robust immune response from the plant, AvrHah1 is not required for these effects. In addition, we demonstrate that cellular leakage that occurs during disease is independent of AvrHah1. Investigation of the interaction between S. enterica, X. gardneri, and the plant host provides information regarding how an inhospitable environment changes during infection and can be transformed into a habitable niche.

|
Scooped by
dromius
January 30, 2022 11:04 AM
|
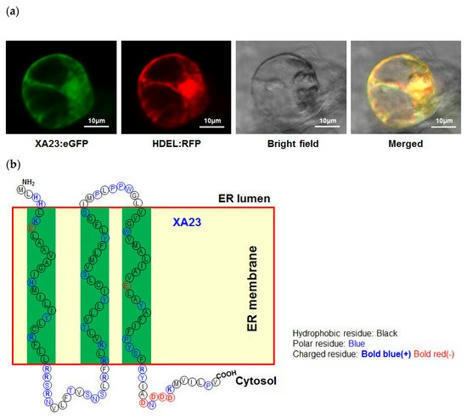
(via T. Lahaye, thx) Xu et al, 2022 - This study, for the first time, uncover a naturally-emerging Xa23-breaking Xoo isolate.
- The ability of AvrXa23 to be trapped by the EBE of Xa23 gene can be altered by one or more RVDs of AvrXa23.
- Seven AvrXa23-like TALEs determine the “arms-race” in the AvrXa23 and Xa23 pair.
- This study provide new insights into the diversified strategies used by Xoo to evade host resistance.
- Planting single R gene (like Xa23) rice in a great region is risk to make Xoo minority to majority.

|
Scooped by
dromius
December 22, 2021 4:52 AM
|
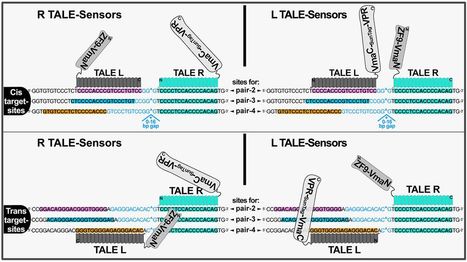
(via T. Schreiber, thx) Taghbalout et al, 2021 Here we describe TALE.Sense, a versatile platform for sensing DNA sequences in live mammalian cells enabling programmable generation of a customable response that discerns cells containing specified sequence targets. The platform is based on the programmable DNA binding of transcription activator-like effector (TALE) coupled to conditional intein-reconstitution producing a trans-spliced ON-switch for a response circuit. TALE.Sense shows higher efficiency and dynamic range when compared to the reported zinc-finger based DNA-sensor in detecting same DNA sequences. Swapping transcriptional activation modules and introducing SunTag-based amplification loops to TALE.Sense circuits augment detection efficiency of the DNA sensor. The TALE.Sense platform shows versatility when applied to a range of target sites, indicating its suitability for applications to identify live cell variants with anticipated DNA sequences. TALE.Sense could be integrated with other cellular or synthetic circuits by using specified DNA sequences as control-switches, thus expanding the scope in connecting inducible modules for synthetic biology.

|
Scooped by
dromius
December 18, 2021 9:49 AM
|
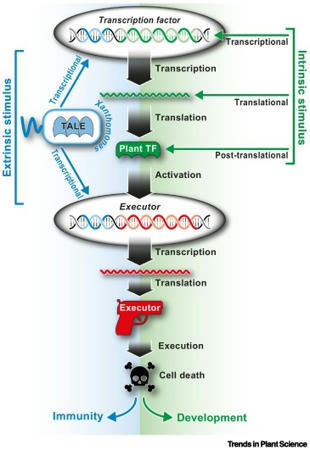
Nowak et al, 2021 Xanthomonads inject transcription-activator like effectors (TALEs) into plant cells, where TALEs bind to effector binding elements (EBEs) in plant promoters and transcriptionally activate downstream host susceptibility genes to promote disease. Some plant genotypes contain transcriptionally activatable cell death-inducing genes, termed executors, that are preceded by TALE-compatible EBEs. Such EBE-equipped executor alleles are, by definition, resistance ( R) genes, since their presence provides protection against TALE-containing xanthomonads. Recent studies uncovered that TALEs can rapidly change their sequence specificity by rearranging their modular DNA domain to circumvent detection by matching executor-type R genes. Evolutionary evidence suggests that the native function of executors is likely to be not in plant immunity, but rather in the regulation of programmed cell death, possibly in the context of plant developmental processes.
|

|
Scooped by
dromius
September 7, 2022 5:12 PM
|
(via T. Lahaye, thx) Cho et al. 2022
Mitochondrial DNA (mtDNA) editing paves the way for disease modeling of mitochondrial genetic disorders in cell lines and animals and also for the treatment of these diseases in the future. Bacterial cytidine deaminase DddA-derived cytosine base editors (DdCBEs) enabling mtDNA editing, however, are largely limited to C-to-T conversions in the 5′-TC context (e.g., TC-to-TT conversions), suitable for generating merely 1/8 of all possible transition (purine-to-purine and pyrimidine-to-pyrimidine) mutations. Here, we present transcription-activator-like effector (TALE)-linked deaminases (TALEDs), composed of custom-designed TALE DNA-binding arrays, a catalytically impaired, full-length DddA variant or split DddA originated from Burkholderia cenocepacia, and an engineered deoxyadenosine deaminase derived from the E. coli TadA protein, which induce targeted A-to-G editing in human mitochondria. Custom-designed TALEDs were highly efficient in human cells, catalyzing A-to-G conversions at a total of 17 target sites in various mitochondrial genes with editing frequencies of up to 49%.

|
Scooped by
dromius
September 3, 2022 11:03 AM
|
Schenstnyi et al. 2022
The Xanthomonas transcription activator-like effector (TALE) protein AvrBs3 transcriptionally activates the executor-type resistance (R) gene Bs3 from pepper (Capsicum annuum), thereby triggering a hypersensitive response (HR). AvrBs3 also triggers HR in tomato (Solanum lycopersicum) upon recognition by the nucleotide-binding leucine-rich repeat (NLR) R protein Bs4. Whether the executor-type R protein Bs3 and the NLR-type R protein Bs4 use common or distinct signalling components to trigger HR remains unclear.
CRISPR/Cas9-mutagenesis revealed, that the immune signalling node EDS1 is required for Bs4- but not for Bs3-dependent HR, suggesting that NLR- and executor-type R proteins trigger HR via distinct signalling pathways. CRISPR/Cas9-mutagenesis also revealed that tomato Bs4 suppresses the virulence function of both TALEs, the HR-inducing AvrBs3 protein and of AvrHah1, a TALE that does not trigger HR in tomato.
Analysis of AvrBs3- and AvrHah1-induced host transcripts and disease phenotypes in CRISPR/Cas9-induced bs4 mutant plants indicates that both TALEs target orthologous transcription factor genes to promote disease in tomato and pepper host plants.
Our studies display that tomato mutants lacking the TALE-sensing Bs4 protein provide a novel platform to either uncover TALE-induced disease phenotypes or genetically dissect components of executor-triggered HR.

|
Scooped by
dromius
July 29, 2022 6:11 PM
|
Lei et al. 2022 DddA-derived cytosine base editors (DdCBEs)—which are fusions of split DddA halves and transcription activator-like effector (TALE) array proteins from bacteria—enable targeted C•G-to-T•A conversions in mitochondrial DNA1. However, their genome-wide specificity is poorly understood. Here we show that the mitochondrial base editor induces extensive off-target editing in the nuclear genome. Genome-wide, unbiased analysis of its editome reveals hundreds of off-target sites that are TALE array sequence (TAS)-dependent or TAS-independent. TAS-dependent off-target sites in the nuclear DNA are often specified by only one of the two TALE repeats, challenging the principle that DdCBEs are guided by paired TALE proteins positioned in close proximity. TAS-independent off-target sites on nuclear DNA are frequently shared among DdCBEs with distinct TALE arrays. Notably, they co-localize strongly with binding sites for the transcription factor CTCF and are enriched in topologically associating domain boundaries. We engineered DdCBE to alleviate such off-target effects. Collectively, our results have implications for the use of DdCBEs in basic research and therapeutic applications, and suggest the need to thoroughly define and evaluate the off-target effects of base-editing tools.

|
Scooped by
dromius
July 7, 2022 5:06 AM
|
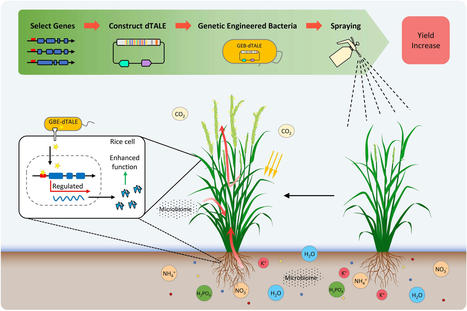
(via T. Schreiber, thx) Tang et al. 2022 Recent studies have shown that reprogramming of gene expression in a genome can induce the production of proteins enabling yield increase. The transcription activator-like effectors (TALEs) from several species of bacterial Xanthomonas have been extensively studied, and a series of research tools, such as genome editing tool TALENs and gene expression activators, have been developed based on the specific protein–nucleic acid recognition and binding mechanisms of TALEs. In this proof-of-principle study, we designed and constructed a designer TALE (dTALE), designated as dTALE-NOG1, to specifically target the promoter of OsNOG1 gene in rice, and demonstrated that this dTALE can be used as a new type of plant growth regulator for better crop growth and harvest. In doing so, the dTALE-NOG1 was transferred into the non-pathogenic Xanthomonas oryzae pv. oryzae (Xoo) strain PH to generate a genetically engineered bacteria (GEB) strain called PH-dtNOG1. Functional verification showed that dTALE-NOG1 could significantly induce the expression of OsNOG1. By spraying cell suspension of PH-dtNOG1 on the rice plants during the tillering stage, the transcription level of OsNOG1 was highly enhanced, the grain number of rice plants was increased by more than 11.40%, and the grain yield per plant increased by more than 11.08%, demonstrating that the dTALE-NOG1 was highly effective in enhancing rice yield. This work provided a new strategy for manipulating agronomical traits by reprogrammin

|
Scooped by
dromius
May 17, 2022 5:09 AM
|
(via T. Schreiber, thx) Doucouré et al, 2022
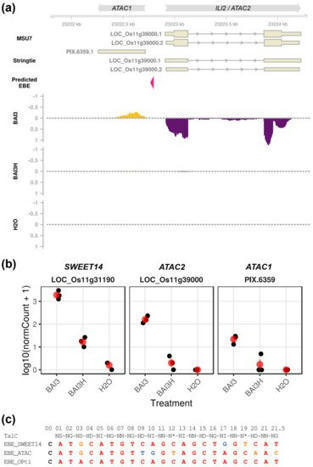
Xanthomonas oryzae pv. oryzae (Xoo) strains that cause bacterial leaf blight (BLB) limit rice (Oryza sativa) production and require breeding more resistant varieties. Transcription activator-like effectors (TALEs) activate transcription to promote leaf colonization by binding to specific plant host DNA sequences termed effector binding elements (EBEs). Xoo major TALEs universally target susceptibility genes of the SWEET transporter family. TALE-unresponsive alleles of clade III OsSWEET susceptibility gene promoter created with genome editing confer broad resistance on Asian Xoo strains. African Xoo strains rely primarily on the major TALE TalC, which targets OsSWEET14. Although the virulence of a talC mutant strain is severely impaired, abrogating OsSWEET14 induction with genome editing does not confer equivalent resistance on African Xoo. To address this contradiction, we postulated the existence of a TalC target susceptibility gene redundant with OsSWEET14. Bioinformatics analysis identified a rice locus named ATAC composed of the INCREASED LEAF INCLINATION 2 (ILI2) gene and a putative lncRNA that are shown to be bidirectionally upregulated in a TalC-dependent fashion. Gain-of-function approaches with designer TALEs inducing ATAC sequences did not complement the virulence of a Xoo strain defective for SWEET gene activation. While editing the TalC EBE at the ATAC loci compromised TalC-mediated induction, multiplex edited lines with mutations at the OsSWEET14 and ATAC loci remained essentially susceptible to African Xoo strains. Overall, this work indicates that ATAC is a probable TalC off-target locus but nonetheless documents the first example of divergent transcription activation by a native TALE during infection.

|
Scooped by
dromius
April 19, 2022 12:25 PM
|
(via T. Lahaye, thx) Mok et al, 2022 The all-protein cytosine base editor DdCBE uses TALE proteins and a double-stranded DNA-specific cytidine deaminase (DddA) to mediate targeted C•G-to-T•A editing. To improve editing efficiency and overcome the strict TC sequence-context constraint of DddA, we used phage-assisted non-continuous and continuous evolution to evolve DddA variants with improved activity and expanded targeting scope. Compared to canonical DdCBEs, base editors with evolved DddA6 improved mitochondrial DNA (mtDNA) editing efficiencies at TC by 3.3-fold on average. DdCBEs containing evolved DddA11 offered a broadened HC (H = A, C or T) sequence compatibility for both mitochondrial and nuclear base editing, increasing average editing efficiencies at AC and CC targets from less than 10% for canonical DdCBE to 15–30% and up to 50% in cell populations sorted to express both halves of DdCBE. We used these evolved DdCBEs to efficiently install disease-associated mtDNA mutations in human cells at non-TC target sites. DddA6 and DddA11 substantially increase the effectiveness and applicability of all-protein base editing.

|
Scooped by
dromius
March 16, 2022 12:16 PM
|
(via T. Schreiber, thx) Sivaram et al, 2022 The molecular mechanism of pomegranate susceptibility to bacterial blight, a serious threat to pomegranate production in India, is largely unknown. In the current study, we have used PacBio and Illumina sequencing of Xanthomonas citri pv. punicae (Xcp) strain 119 genome to identify tal genes and RNA-Seq analysis to identify putative host targets in the susceptible pomegranate variety Bhagwa challenged with Xcp119. Xcp119 genome encodes seven transcription activator-like effectors (TALEs), three of which are harbored by a plasmid. RVD-based phylogenetic analysis of TALEs of Xanthomonas citri pathovars indicate the TALEs of Xcp as evolutionarily and functionally close to Xanthomonas citri pv. malvacearum and Xanthomonas citri pv. glycines. Comparative RNA-Seq of Xcp and mock-inoculated leaf tissues revealed Xcp-induced pomegranate transcription modulation. The prediction of TALE binding elements (EBEs) in the promoters of up-regulated genes identified a set of TALE-targeted candidate genes in pomegranate–Xcp interaction. The predicted candidate susceptibility genes include two oxoglutarate-dependent dioxygenase gene, ethylene-responsive transcription factor and flavanone 3-hydroxylase-like gene, and the further characterization of these would enable blight resistance engineering in pomegranate.

|
Scooped by
dromius
March 6, 2022 2:56 PM
|
(via T. Schreiber, thx) Raeisi et al, 2022
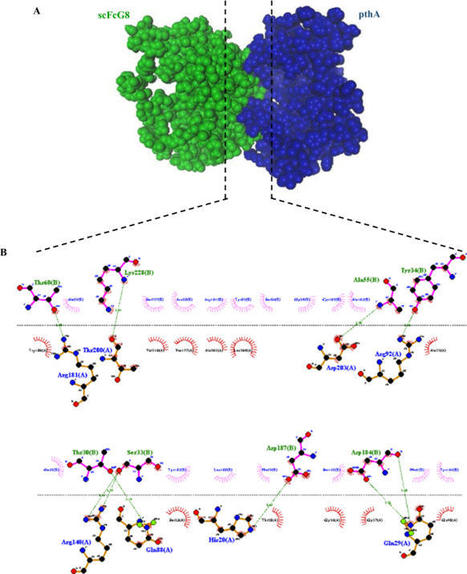
Citrus bacterial canker, caused by Xanthomonas citri subsp. citri (Xcc), is a major disease of citrus plants, causing a significant loss in the citrus industry. The pthA is a bacterial effector protein mediates protein–protein and protein-DNA interactions and modulates host transcription. Injection of pthA effector protein into the host cell induces the expression of the susceptibility gene CsLOB1 which is required for citrus canker disease development. In this study, we described in planta expression of a specific anti-pthA single-chain variable fragment (scFv) recombinant antibody, scFvG8, and assessed its function using molecular docking, immunoblotting, and indirect enzyme-linked immunosorbent assay (ELISA). Based on the results, homology-based molecular docking suggested that at least eight intermolecular hydrogen bonds are involved in pthA-scFvG8 interactions. Immunoblotting and indirect ELISA results reconfirmed specific binding of scFvG8 to pthA protein. Moreover, gene fragment encoding scFvG8 was cloned into plant expression vector and transiently expressed in leaves of Nicotiana tabacum cv. Samson by agroinfiltration method. Transient expression of scFvG8 (at the expected size of 35 kDa) in N. tabacum leaves was confirmed by western blotting. Also, immunoblotting and indirect ELISA showed that the plant-derived scFvG8 had similar activity to purified scFvG8 antibody in detecting pthA. Additionally, in scFvG8-expressing tobacco leaves challenged with Xcc, a reduction (for up to 70%) of hypersensitive response (HR) possibly via direct interaction with pthA, was observed in the necrotic leaf area compared to control plants infected with empty vector. The results obtained in this study confirm that scFvG8 can suppress the function of pthA effector protein within plant cells, thus the induction of stable expression of scFvG8 in lime trees can be considered as an appropriate approach to confer resistance to Xcc.

|
Scooped by
dromius
February 6, 2022 8:03 AM
|
Ji and Guo et al, 2022 Executor (E) genes comprise a new type of plant resistance (R) genes, identified from host–Xanthomonas interactions. The Xanthomonas-secreted transcription activation-like effectors (TALEs) usually function as major virulence factors, which activate the expression of the so-called “susceptibility” (S) genes for disease development. This activation is achieved via the binding of the TALEs to the effector-binding element (EBE) in the S gene promoter. However, host plants have evolved EBEs in the promoters of some otherwise silent R genes, whose expression directly causes a host cell death that is characterized by a hypersensitive response (HR). Such R genes are called E genes because they trap the pathogen TALEs in order to activate expression, and the resulting HR prevents pathogen growth and disease development. Currently, deploying E gene resistance is becoming a major component in disease resistance breeding, especially for rice bacterial blight resistance. Currently, the biochemical mechanisms, or the working pathways of the E proteins, are still fuzzy. There is no significant nucleotide sequence homology among E genes, although E proteins share some structural motifs that are probably associated with the signal transduction in the effector-triggered immunity. Here, we summarize the current knowledge regarding TALE-type avirulence proteins, E gene activation, the E protein structural traits, and the classification of E genes, in order to sharpen our understanding of the plant E genes.

|
Scooped by
dromius
February 1, 2022 5:27 AM
|
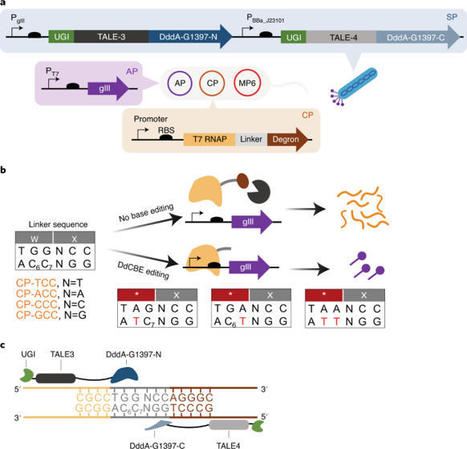
(via T. Schreiber, thx) Wei et al, 2022 Although ZFN and TALEN have been previously engineered to successfully cut and eliminate mtDNA in a programmable way5,6,7,8,9, correction of disease-causing point mutations in mtDNA remained challenging. Recently, DddA-derived cytosine base editors (DdCBEs) have been developed to specifically induce C-to-T conversion in mtDNA by fusion of sequence-programmable TALE and split deaminase derived from interbacterial toxins10.
In this study, we sought to test DdCBE’s ability for base editing of mtDNA in human embryos.
Taken together, our results demonstrate that DdCBE is an effective base editor for inducing point mutations in mtDNA of human embryos, and the efficiency is much higher in 8-cell embryos. Our 8-cell injection method could help generate mitochondrial disease models as well as derived embryonic stem cells for functional investigation of disease-associated mutations in mtDNA.

|
Scooped by
dromius
January 18, 2022 8:22 AM
|
Liu, et al 2021
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. Another drawback of TALEs is their tendency to bind and cleave off-target sequence, which hampers their clinical application and renders applications requiring high-fidelity binding unfeasible. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and/or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.

|
Scooped by
dromius
December 21, 2021 3:56 AM
|
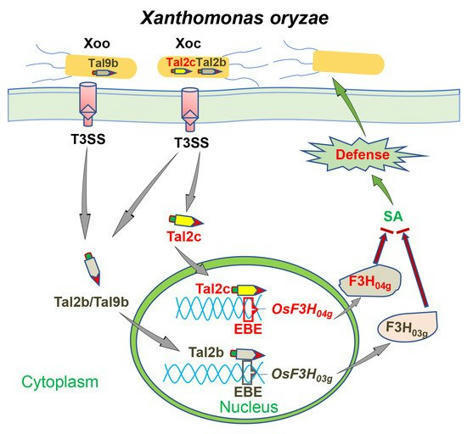
(via T. Schreiber, thx) Wu et al, 2021 Xanthomonas oryzae delivers transcription activator-like effectors (TALEs) into plant cells to facilitate infection. Following economic principles, the redundant TALEs are rarely identified in Xanthomonas. Previously, we identified the Tal2b, which activates the expression of the rice 2-oxoglutarate-dependent dioxygenase gene OsF3H03g to promote infection in the highly virulent strain of X. oryzae pv. oryzicola HGA4. Here, we reveal that another clustered TALE, Tal2c, also functioned as a virulence factor to target rice OsF3H04g, a homologue of OsF3H03g. Transferring Tal2c into RS105 induced expression of OsF3H04g to coincide with increased susceptibility in rice. Overexpressing OsF3H04g caused higher susceptibility and less salicylic acid (SA) production compared to wild-type plants. Moreover, CRISPR–Cas9 system-mediated editing of the effector-binding element in the promoters of OsF3H03g or OsF3H04g was found to specifically enhance resistance to Tal2b- or Tal2c-transferring strains, but had no effect on resistance to either RS105 or HGA4. Furthermore, transcriptome analysis revealed that several reported SA-related and defense-related genes commonly altered expression in OsF3H04g overexpression line compared with those identified in OsF3H03g overexpression line. Overall, our results reveal a functional redundancy mechanism of pathogenic virulence in Xoc in which tandem Tal2b and Tal2c specifically target homologues of host genes to interfere with rice immunity by reducing SA.
|



 Your new post is loading...
Your new post is loading...