Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
April 16, 2012 3:34 PM
|
The Two Blades Foundation (2Blades) has completed a non-exclusive license agreement with the Monsanto Company for access to the TAL Code technology for genome engineering in plants. The Transcription Activator Like (TAL) effector Code technology, discovered by Ulla Bonas, Jens Boch, Thomas Lahaye, and Sebastian Schornack at Martin-Luther University in Halle, Germany, is based on novel sequence-specific DNA-binding proteins that can be designed quickly and easily to recognize virtually any sequence of interest. Named Method of the Year in 2011 by the journal Nature Methods (9:1 doi:10.1038/nmeth.1852), the technology enables a number of highly useful tools to target specific loci in a genome and modulate the expression of genes. The application of these tools in plants will accelerate improvements in crop growth and development. The Two Blades Foundation holds exclusive rights for commercial uses of the technology in plants. "Having Monsanto, the world's largest seed company, put the TAL Code technology to use in their programs will further ensure its wide use and development," said 2Blades Chief Operating Officer Diana Horvath. 2Blades will gain access to Monsanto's improvements to the technology for use in 2Blades' humanitarian efforts in support of subsistence farming. The license agreement will aid Monsanto's mission to develop high quality products for sustainable agriculture through science-based solutions. Financial terms of the agreement were not disclosed.

|
Scooped by
dromius
April 10, 2012 5:22 PM
|
Nature Biotech - FLASH assembly of TALENs for high-throughput genome editing
Reyon et al. (2012) http://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.2170.html Engineered transcription activator–like effector nucleases (TALENs) have shown promise as facile and broadly applicable genome editing tools. However, no publicly available high-throughput method for constructing TALENs has been published, and large-scale assessments of the success rate and targeting range of the technology remain lacking. Here we describe the fast ligation-based automatable solid-phase high-throughput (FLASH) system, a rapid and cost-effective method for large-scale assembly of TALENs. We tested 48 FLASH-assembled TALEN pairs in a human cell–based EGFP reporter system and found that all 48 possessed efficient gene-modification activities. We also used FLASH to assemble TALENs for 96 endogenous human genes implicated in cancer and/or epigenetic regulation and found that 84 pairs were able to efficiently introduce targeted alterations. Our results establish the robustness of TALEN technology and demonstrate that FLASH facilitates high-throughput genome editing at a scale not currently possible with other genome modification technologies.

|
Scooped by
dromius
April 10, 2012 4:14 AM
|
The method developed by Joung and his colleagues – called the FLASH (fast ligation-based automatable solid-phase high-throughput) system – assembles DNA fragments encoding a TALEN on a magnetic bead held in place by an external magnet, allowing automated construction by a liquid-handling robot of DNA that encodes as many as 96 TALENs in a single day at a cost of around $75 per TALEN. Joung's team also developed a manual version of FLASH that would allow labs without access to robotic equipment to construct up to 24 TALEN sequences a day. In their test of the system in human cells, the investigators found that FLASH-assembled TALENs were able to successfully induce breaks in 84 of 96 targeted genes known to be involved in cancer or in epigenetic regulation.

|
Scooped by
dromius
March 23, 2012 6:08 PM
|
Nature Methods - How TAL effectors bind DNA
Monya Baker highlights two publications about TAL effector repeat domain. A right handed super helix wraps itself along the major groove of DNA...One of the two variable amino acids makes a specific contact with a nucleotide in the DNA sense strand while the other stabilizes the contact between DNA and protein.

|
Suggested by
Tom Ellis
March 22, 2012 6:47 AM
|
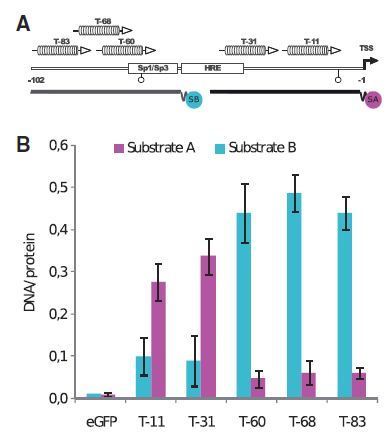
Transcription Activator-Like Orthogonal Repressors (TALORs) are a new tool for scalable designer regulation of synthetic promoters. TALORS are a modification to customisable Transcription Activator-Like Effectors (TALEs) that binds core promoter regions to act as transcriptional repressors.

|
Scooped by
dromius
March 13, 2012 5:24 PM
|
Bradley - 2012 Here we describe a structure prediction protocol tailored to the TAL–DNA system, and report simulation results that shed light on observed repeat-base associations and overall TAL structure. Our models demonstrate that TAL–DNA interactions can be explained by a model in which the TAL repeat domain forms a superhelical repeat structure that wraps around undistorted B-form DNA, paralleling the geometry of the major groove, with contacts between position 13 of each repeat and its associated base pair on the sense strand determining the specificity of DNA recognition.

|
Scooped by
dromius
March 2, 2012 6:30 AM
|
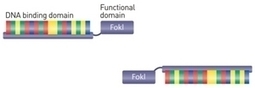
As DNA molecule is so extensively characterized, being sort of a pop culture molecule, finding a completely novel way of using it presents quite a challenge. The main twist comes with the requirement that each of those DNA-binding proteins is fused to a different functional protein. Therefore, the sequence of target motifs encoded by DNA program defines also the arrangement of those functional proteins along with the order of DNA-binding factors. What is so powerful about this idea is the fact that by only changing the sequence of a DNA program, either switching positions or adding new target sequences, the outcome can be predicted in advance. The first big application is to use the DNA program to arrange the sequence of biosynthetic reaction enzymes and therefore guide the biosynthetic flow towards the desired products. the flow of information can be restricted to the reactions occurring along the linear DNA instead of throughout the cell. Attachment of the cascade of proteases, phosphatases or other enzymes/protein interaction domains to the DNA scaffold could result in a rapid information processing depending on the input of particular DNA sequence and initiation reaction. use DNA-binding factors to extend and simplify the construction of oscillators.

|
Scooped by
dromius
February 5, 2012 4:27 PM
|
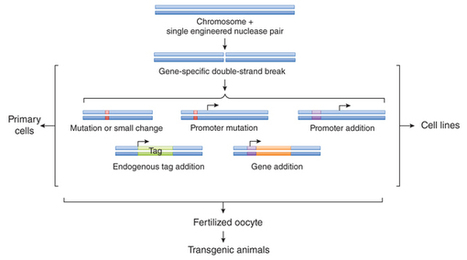
Ning Sun , Jing Liang , Zhanar Abil and Huimin Zhao; Mol. BioSyst., 2012, Advance Article TAL effector nucleases (TALENs) represent a new class of artificial nucleases capable of cleaving long, specific target DNA sequences in vivo and are powerful tools for genome editing with potential therapeutic applications. Here we report a pair of custom-designed TALENs for targeted genetic correction of the sickle cell disease mutation in human cells, which represents an example of engineered TALENs capable of recognizing and cleaving a human disease-associated gene. A systematic study was carried out to optimize TALEN architecture for maximal in vivo cleavage efficiency. In contrast to the previous reports, the engineered TALENs were capable of recognizing and cleaving target binding sites preceded by A, C or G. More importantly, the optimized TALENs efficiently cleaved a target sequence within the human β-globin (HBB) gene associated with sickle cell disease and increased the efficiency of targeted gene repair by >1000-fold in human cells. In addition, these TALENs showed no detectable cytotoxicity.

|
Scooped by
dromius
January 28, 2012 11:30 AM
|
from the lab of Magdy M. Mahfouz: "Here, we report the development and use of a rapid and straightforward approach for the construction of designer TALE (dTALE) activators and nucleases with user-selected DNA target specificity. Using our plasmid set of 100 repeat modules, researchers can assemble repeat domains for any 14-nucleotide target sequence in one sequential restriction-ligation cloning step and in only 24 h. We generated several custom dTALEs and dTALENs with new target sequence specificities and validated their function by transient expression in tobacco leaves and in vitro DNA cleavage assays, respectively. Our dTALE repeat assembly approach along with the web tool idTALE will expedite genome-engineering applications in a variety of cell types and organisms including plants."

|
Scooped by
dromius
January 17, 2012 4:29 AM
|
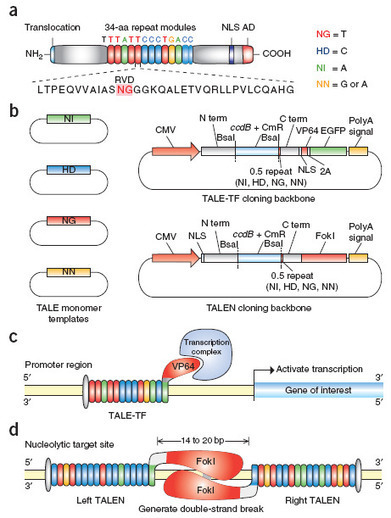
A transcription activator-like effector toolbox for genome engineering - Nature Protocols
Sanjana et al, Nature Protocols7,171–192(2012) Transcription activator-like effectors (TALEs) are a class of naturally occurring DNA-binding proteins found in the plant pathogen Xanthomonas sp. The DNA-binding domain of each TALE consists of tandem 34–amino acid repeat modules that can be rearranged according to a simple cipher to target new DNA sequences. Customized TALEs can be used for a wide variety of genome engineering applications, including transcriptional modulation and genome editing. Here we describe a toolbox for rapid construction of custom TALE transcription factors (TALE-TFs) and nucleases (TALENs) using a hierarchical ligation procedure. This toolbox facilitates affordable and rapid construction of custom TALE-TFs and TALENs within 1 week and can be easily scaled up to construct TALEs for multiple targets in parallel. We also provide details for testing the activity in mammalian cells of custom TALE-TFs and TALENs using quantitative reverse-transcription PCR and Surveyor nuclease, respectively. The TALE toolbox described here will enable a broad range of biological applications.

|
Scooped by
dromius
January 5, 2012 5:59 PM
|
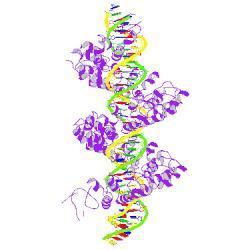
Each repeat forms a left-handed, two-helix bundle that presents an RVD-containing loop to the DNA. The repeats self-associate to form a right-handed superhelix wrapped around the DNA major groove. Two degenerate N-terminal repeats also interact with the DNA. Figure: Fig.3, Amanda Nga-Sze Mak et al. (2012)

|
Scooped by
dromius
January 5, 2012 10:04 AM
|
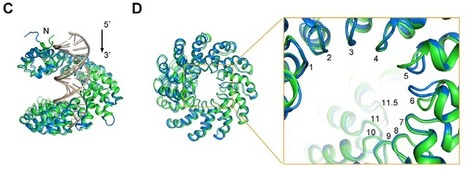
Mak, A.N.S., Bradley, P., Cernadas, R.A., Bogdanove, A.J., Stoddard, B.L., Journal: (2012) Science source: pdb database (rcsb.org)

|
Scooped by
dromius
December 29, 2011 3:09 PM
|
Engineered nucleases have advanced the field of gene therapy with the promise of targeted genome modification as a treatment for human diseases.
|

|
Scooped by
dromius
April 14, 2012 2:07 PM
|
Bultmann et al (2012) Specific control of gene activity is a valuable tool to study and engineer cellular functions. Recent studies uncovered the potential of transcription activator-like effector (TALE) proteins that can be tailored to activate user-defined target genes. It remains however unclear whether and how epigenetic modifications interfere with TALE-mediated transcriptional activation. We studied the activity of five designer TALEs (dTALEs) targeting the oct4 pluripotency gene. In vitro assays showed that the five dTALEs that target distinct sites in the oct4 promoter had the expected DNA specificity and comparable affinities to their corresponding DNA targets. In contrast to their similar in vitro properties, transcriptional activation of oct4 by these distinct dTALEs varied up to 25-fold. While dTALEs efficiently upregulated transcription of the active oct4 promoter in embryonic stem cells (ESCs) they failed to activate the silenced oct4 promoter in ESC-derived neural stem cells (NSCs), indicating that as for endogenous transcription factors also dTALE activity is limited by repressive epigenetic mechanisms. We therefore targeted the activity of epigenetic modulators and found that chemical inhibition of histone deacetylases by valproic acid or DNA methyltransferases by 5-aza-2′-deoxycytidine facilitated dTALE-mediated activation of the epigenetically silenced oct4 promoter in NSCs. Notably, demethylation of the oct4 promoter occurred only if chemical inhibitors and dTALEs were applied together but not upon treatment with inhibitors or dTALEs only. These results show that dTALEs in combination with chemical manipulation of epigenetic modifiers facilitate targeted transcriptional activation of epigenetically silenced target genes.

|
Scooped by
dromius
April 10, 2012 5:18 PM
|
FLASH is an automatable high-throughput method for assembling DNA encoding TAL effector repeat arrays recently developed by the Joung lab (Reyon & Tsai et al., Nat Biotechnol. 2012). With automated FLASH, DNA fragments encoding 96 variable-length TAL effector repeat arrays can be assembled in one day. When practiced manually with a multi-channel pipet, FLASH can also be used by a single researcher to make DNA fragments encoding 12 to 24 variable-length TAL effector repeat arrays in one to two days. TAL effector repeat arrays assembled by FLASH are identical in DNA sequence to those assembled by the REAL and REAL-Fast methods.

|
Scooped by
dromius
March 28, 2012 5:45 AM
|
http://idtale.kaust.edu.sa/ idTALE is a web-based tool developed to facilitate TALEN design and target finding in the genomes of several model species. idTALE helps in TALEN design as well as to search for its targets in the genomes of multiple model organisms.

|
Scooped by
dromius
March 22, 2012 7:17 AM
|
“What this technology allows you to do is to make direct edits to the genome,” said Reed Hickey, Life Technologies’ product manager. Data gathered by the company have proved its effectiveness with fungi, algae and a variety of other biomass forms. “What this allows you to do is turn on a whole pathway, [for example to] increase the expression of lipid production in a specific strain of algae, or change the way photosynthesis is performed so you can support photosynthetic pathways from other strains and engineer a plant to do exactly what you need it to do to improve the energy capture for biofuels.”

|
Scooped by
dromius
March 19, 2012 5:40 AM
|
GeneArt® Precision TALs provide custom DNA binding proteins for accurate DNA targeting and precise genome editing.

|
Scooped by
dromius
March 3, 2012 12:49 AM
|
de Souza et al, 2012, PLoS One Although target genes and DNA specificity of TAL effectors have been elucidated, how TAL proteins control host transcription is poorly understood.... To extend our knowledge on the mode of action of PthAs, we have identified new protein targets of the PthA4 variant, required to elicit canker on citrus. Here we show that all the PthA4-interacting proteins are DNA and/or RNA-binding factors implicated in chromatin remodeling and repair, gene regulation and mRNA stabilization/modification. The majority of these proteins...interacted with each other, suggesting that they assemble into a multiprotein complex. CsHMG was shown to bind DNA and to interact with the invariable leucine-rich repeat region of PthAs. Surprisingly, both CsHMG and PthA4 interacted with PABP1 and 2 and showed selective binding to poly(U) RNA, a property that is novel among HMGs and TAL effectors. Given that homologs of CsHMG, CsPABP1, CsPABP2, CsSMC and CsTRAX in other organisms assemble into protein complexes to regulate mRNA stability and translation, we suggest a novel role of TAL effectors in mRNA processing and translational control.

|
Scooped by
dromius
February 5, 2012 4:48 PM
|
Aleksandra Filipovska and Oliver Rackham Mol. BioSyst., 2012 Sequence specific binding of DNA and RNA is of fundamental importance in the regulation of cellular gene expression. Because of their modular structure repeat domain proteins are particularly well suited for these processes and have been widely adopted throughout evolution. Detailed biochemical and structural data has revealed the key residues responsible for recognition of RNA by Pumilio and FBF homology (PUF) repeat proteins and shown that the base specificity can be predicted and re-engineered. Recent work on the DNA-binding properties of transcription activator-like effector (TALE) proteins has shown that their specificity also relies on only a few key residues with a predictable code that can be used to design new DNA-binding proteins. Although less well understood, pentatricopeptide repeat (PPR) proteins contain motifs that appear to contribute to RNA recognition and comparisons to TALE and PUF proteins may help elucidate the code by which they recognize their RNA targets. Understanding how repeat proteins bind nucleic acids enables their biological roles to be uncovered and the design of engineered proteins with predictable RNA and DNA targets for use in biotechnology.

|
Scooped by
dromius
February 1, 2012 5:25 PM
|
The TAL family of bacterial DNA-binding proteins is perhaps the most amazing example of modular DNA recognition imaginable, eclipsing even the ubiquitous zinc finger in terms of beauty and simplicity.

|
Scooped by
dromius
January 17, 2012 4:11 PM
|
EVANSTON, IL (January 16, 2012) -- The Two Blades Foundation (2Blades) announced today the completion of a non-exclusive license agreement with Syngenta, which provides Syngenta with access to TAL Code technology for commercial uses in certain crop plants.

|
Scooped by
dromius
January 5, 2012 6:16 PM
|
Researchers at Fred Hutchinson Cancer Research Center have solved the three-dimensional structure of a newly discovered type of gene-targeting protein that has shown to be useful as a DNA-targeting molecule for gene correction, gene therapy and...

|
Scooped by
dromius
January 5, 2012 5:58 PM
|
Here, we report the crystal structures of a 11.5-repeat TAL effector in both DNA-free and DNA-bound states. Each TAL repeat comprises two helices connected by a short RVD-containing loop. The 11.5 repeats form a right-handed, super-helical structure that tracks along the sense strand of DNA duplex, with RVDs contacting the major groove. The 12th residue stabilizes the RVD loop, whereas the 13th residue makes a base-specific contact. Understanding DNA recognition by TAL effectors may facilitate rational design of DNA-binding proteins with biotechnological applications. Fig.: suppl. Figure S3, Dong Deng et al, 2012

|
Scooped by
dromius
January 5, 2012 1:46 AM
|
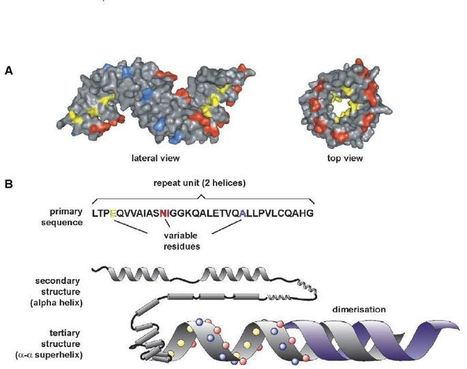
AvrBs3-like proteins are predicted to form a helical superstructure that resembles a tetratricopeptide repeat (TPR) fold. Variable repeat unit residues 4 (yellow), 12+13 (red) and 24 (blue) are depicted. (A) 3D Jury/MODELLER surface model was predicted based on the crystal structure of the TPR domain of the O-linked GLCNAC transferase (Protein Database (PDB) entry: 1w3b_A) and is displayed as lateral and top view. (B) Schematic illustration of the predicted right-handed α–α helical superstructure of AvrBs4 and its structural hierarchy. A second AvrBs3-like protein (blue) illustrates the postulated dimerization of this protein type.
|



 Your new post is loading...
Your new post is loading...