Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
July 10, 2012 8:36 PM
|
TAL effector RVD specificities and efficiencies - Nature Biotech
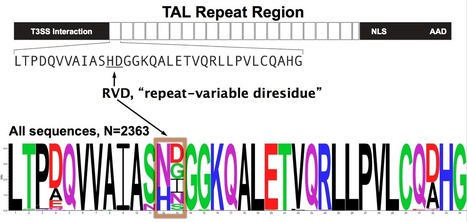
http://www.nature.com/nbt/journal/v30/n7/full/nbt.2304.html The DNA-binding domain of transcription activator–like effectors (TALEs) has become an important tool for the programmable and specific targeting of genome-editing nucleases. We determined specificities of TALE DNA-binding modules and discovered varying efficiencies to promote TALE activity. Specific recognition of guanine is difficult because the common RVD NN (Asn-Asn) recognizes guanine and adenine, whereas the guanine-specific RVD NK (Asn-Lys) apparently functions less well than NN does11. Here we describe new RVD specificities including NH (Asn-His), which is highly specific for guanine. We show that efficiency of RVDs varies and that strong repeats (HD (His-Asn) or NN) are key for overall activity of TALEs.

|
Scooped by
dromius
July 10, 2012 3:25 AM
|
In order to promote virulence, Gram-negative bacteria have evolved the ability to inject so-called type III effector proteins into host cells. ... Mounting evidence suggests that manipulation of host transcription is a major strategy developed by bacteria to counteract plant defense responses. It has been suggested that bacterial effectors may adopt at least three alternative, although not mutually exclusive, strategies to subvert host transcription. T3Es may (1) act as transcription factors that directly activate transcription in host cells, (2) affect histone packing and chromatin configuration, and/or (3) directly target host transcription factor activity. Here, we provide an overview on how all these strategies may lead to host transcriptional re-programming and, as a result, to improved bacterial multiplication inside plant cells.

|
Scooped by
dromius
July 7, 2012 1:42 PM
|
Technology development has always been one of the forces driving breakthroughs in biomedical research. Since the time of Thomas Morgan, Drosophilists have, step by step, developed powerful genetic tools for manipulating and functionally dissecting the Drosophila genome, but room for improving these technologies and developing new techniques is still large, especially today as biologists start to study systematically the functional genomics of different model organisms, including humans, in a high-throughput manner. Here, we report, for the first time in Drosophila, a rapid, easy, and highly specific method for modifying the Drosophila genome at a very high efficiency by means of an improved transcription activator-like effector nuclease (TALEN) strategy. We took advantage of the very recently developed “unit assembly” strategy to assemble two pairs of specific TALENs designed to modify the yellow gene (on the sex chromosome) and a novel autosomal gene. The mRNAs of TALENs were subsequently injected into Drosophila embryos. From 31.2% of the injected F0 fertile flies, we detected inheritable modification involving the yellow gene. The entire process from construction of specific TALENs to detection of inheritable modifications can be accomplished within one month. The potential applications of this TALEN-mediated genome modification method in Drosophila are discussed.

|
Suggested by
Nicolas Denancé
July 3, 2012 12:41 PM
|
Hummel et al (2012), New Phytologist Xanthomonas transcription activator-like (TAL) effectors promote disease in plants by binding to and activating host susceptibility genes. Plants counter with TAL effector-activated executor resistance genes, which cause host cell death and block disease progression. We asked whether the functional specificity of an executor gene could be broadened by adding different TAL effector binding elements (EBEs) to it. We added six EBEs to the rice Xa27 gene, which confers resistance to strains of the bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Xoo) that deliver the TAL effector AvrXa27. The EBEs correspond to three other effectors from Xoo strain PXO99A and three from strain BLS256 of the bacterial leaf streak pathogen Xanthomonas oryzae pv. oryzicola (Xoc).Stable integration into rice produced healthy lines exhibiting gene activation by each TAL effector, and resistance to PXO99A, a PXO99A derivative lacking AvrXa27, and BLS256, as well as two other Xoo and 10 Xoc strains virulent toward wildtype Xa27 plants. Transcripts initiated primarily at a common site. Sequences in the EBEs were found to occur nonrandomly in rice promoters, suggesting an overlap with endogenous regulatory sequences. Thus, executor gene specificity can be broadened by adding EBEs, but caution is warranted because of the possible coincident introduction of endogenous regulatory elements.

|
Scooped by
dromius
June 29, 2012 2:55 AM
|
Cassava bacterial blight (CBB), incited by Xanthomonas axonopodis pv. manihotis (Xam), is the most important bacterial disease of cassava, a staple food source for millions of people in developing countries. [Xam carries several TAL effector genes] that are inaccessible to assembly using
short-read technology because of this highly repetitive central domain and consequently were not revealed during our Illumina assemblies and effector prediction. To gain additional information about Xam TAL effectors, we conducted Southern blot
analysis to estimate the approximate copy number of TAL effector proteins across our Xam isolate collection ... All Xam strains tested contain at least one TAL effector sequence and most strains contain multiple copies. Studies in several other Xanthomonas spp. suggest that TAL effectors play a major role in symptom development. Correspondingly, we found a moderate correlation (R2 = 0.64) between symptom severity and tal gene copy number. However, bacterial growth was only weakly correlated with tal gene copy number, suggesting that TAL effectors may have unequal contributions to overall virulence

|
Scooped by
dromius
May 22, 2012 5:08 AM
|
"Using the TAL effectors to remove DNA will likely challenge anti-GMO opponents. However, since many are also anti-technology, it’s unlikely they will embrace this methodology either."

|
Scooped by
dromius
May 20, 2012 5:38 AM
|
[Zebrafish. 2011] Genome editing appears poised to enter an exciting new era. Targeted double-stranded breaks due to custom restriction enzymes are powerful nucleating events for the induction of local changes in the genome. The zinc finger nuclease (ZFN) platform established the potential of this approach for the zebrafish, but access to high quality reagents has been a major bottleneck for the field. However, two groups recently report successful somatic and germline gene modification using a new nuclease architecture, transcription activator-like effector nucleases (TALENs). TALEN construction is simpler, potentially more reliable, and in the few cases examined, shows fewer off-target effects than corresponding ZFNs. TALENs promise to bring gene targeting to the majority of zebrafish laboratories.

|
Scooped by
dromius
May 16, 2012 6:22 PM
|
Friedreichs ataxia inherited disease (GAA repeat length polymorphism leading to inefficient splicing) http://en.wikipedia.org/wiki/Friedreich%27s_ataxia Genes coding for Tal effector (TALE) proteins may be engineered to target specific DNA sequences. TALEs are fused with a transcription activator can be used to specifically induce the expression of a gene. This could lead to completely new therapies for several diseases. We have applied this potential therapeutic approach to Friedreich ataxia (FRDA) as an example. FRDA is due to a reduced expression of frataxin following an elongation of a trinucleotide (GAA) repeat in intron 1. Aim: To develop a potential treatment for FRDA by increasing the expression of the frataxin gene. Results: We have engineered 12 TALE genes (TALEFrat) coding for TALEFrat proteins each specifically targeting different 14 bp DNA sequences within the proximal region of the human frataxin promoter. When the genes of these TALEFrat were fused with a transcription activator, i.e., four VP16 peptides (i.e., VP64), the resulting TALEFrat-VP64 proteins induced the expression of a mCherry reporter gene fused to a mini-CMV promoter able to be activated by the insertion of the frataxin proximal promoter upstream to the mini promoter. These TALEFrat-VP64 also increased by 2 to 3 folds the frataxin gene expression (detected by qRT-PCR) in the cells. Conclusion: TALEFrat proteins targeting the frataxin promoter are thus a method to increase the expression of frataxin mRNA and potentially could alleviate the symptoms of Friedreich ataxia. This new methodology of TALE effector opens a new field, which could be used to develop TALE proteins to treat other diseases by inducing the expression of specific genes.

|
Scooped by
dromius
May 16, 2012 2:58 AM
|
contains a TAL effector: http://www.ncbi.nlm.nih.gov/protein/CCG39214.1 The 5.1-Mb genome sequence of Xanthomonas citri pv. mangiferaeindicae strain LMG 941, the causal agent of bacterial black spot in mango. Apart from evolutionary studies, the draft genome will be a valuable resource for the epidemiological studies and quarantine of this phytopathogen.

|
Scooped by
dromius
May 13, 2012 12:12 PM
|
Site-specific and adaptable DNA binding domains are essential modules to develop genome engineering technologies for crop improvement. Transcription activator-like effectors (TALEs)
proteins are used to provide highly specific and adaptable DNA binding modules. TALE chimeric nucleases (TALENs) were used to generate site-specific double strand breaks (DSBs) in
vitro and in yeast, Caenorhabditis elegans, mammalian and plant cells. The genomic DSBs can be generated at predefined and user-selected loci and repaired by either non-homologous end joining (NHEJ) or homology dependent repair (HDR). Thus, TALENs can be used to achieve site-specific gene addition, stacking, deletion or inactivation. TALE-based genome engineering tools should be powerful new agricultural biotechnology approaches for crop improvement. Here, we discuss the recent research and the potential applications of TALENs to accelerate the generation of genomic variants through targeted mutagenesis and to produce a nontransgenic GM crops with the desired phenotype.

|
Scooped by
dromius
May 10, 2012 2:36 AM
|
Targeted manipulation of complex genomes often requires the introduction of a double-strand break at defined locations by site-specific DNA endonucleases. Here, we describe a monomeric nuclease domain derived from GIY-YIG homing endonucleases for genome-editing applications. Fusion of the GIY-YIG nuclease domain to three-member zinc-finger DNA binding domains generated chimeric GIY-zinc finger endonucleases (GIY-ZFEs). Significantly, the I-TevI-derived fusions (Tev-ZFEs) function in vitro as monomers to introduce a double-strand break, and discriminate in vitro and in bacterial and yeast assays against substrates lacking a preferred 5'-CNNNG-3' cleavage motif. The Tev-ZFEs function to induce recombination in a yeast-based assay with activity on par with a homodimeric Zif268 zinc-finger nuclease. We also fused the I-TevI nuclease domain to a catalytically inactive LADGLIDADG homing endonuclease (LHE) scaffold. The monomeric Tev-LHEs are active in vivo and similarly discriminate against substrates lacking the 5'-CNNNG-3' motif. The monomeric Tev-ZFEs and Tev-LHEs are distinct from the FokI-derived zinc-finger nuclease and TAL effector nuclease platforms as the GIY-YIG domain alleviates the requirement to design two nuclease fusions to target a given sequence, highlighting the diversity of nuclease domains with distinctive biochemical properties suitable for genome-editing applications.

|
Scooped by
dromius
May 1, 2012 12:11 PM
|
EVANSTON, Ill., May 1, 2012 /PRNewswire-iReach/ -- The Two Blades Foundation (2Blades) announced today the completion of a non-exclusive license agreement with Bayer CropScience AG for access to the TAL Code technology for commercial uses in certain crop plants.

|
Scooped by
dromius
April 17, 2012 1:05 PM
|
Tzfira et al, 2012 Genome editing, i.e. the ability to mutagenize, insert, delete and replace sequences, in living cells is a powerful and highly desirable method that could potentially revolutionize plant basic research and applied biotechnology. Indeed, various research groups from academia and industry are in a race to devise methods and develop tools that will enable not only site-specific mutagenesis but also controlled foreign DNA integration and replacement of native and transgene sequences by foreign DNA, in living plant cells.... In this review, we discuss the principles and tools for restriction enzyme-mediated gene targeting in plant cells, as well as their current and prospective use for gene targeting in model and crop plants.
|

|
Scooped by
dromius
July 10, 2012 12:00 PM
|
The successful candidate will develop site-specific multigene engineering technology in indica rice using the TAL (transcription activator-like) Effector Nucleases and other approaches at IRRI, applicable to assemble drought tolerance genes and C4 candidate gene,working closely with the collaborators under GRiSP new frontier project. He/She will conduct the site specific integration and multi gene engineering, perform molecular analysis and inheritance study in the transgenic progenies.

|
Scooped by
dromius
July 10, 2012 3:10 AM
|
Two Blades Foundation (2Blades) has completed a non-exclusive license agreement with KWS SAAT AG (KWS) for access to 2Blades' Transcription Activator Like (TAL) effector code technology for genome engineering in certain crops.

|
Suggested by
Eli
July 6, 2012 12:26 PM
|
"The TAL Sequence Assembly Tool helps researchers assemble TALE and TALEN plasmids by generating templates and aligning sequencing reads. Compatible with the Golden Gate assembly processes described by Cermak et al. and Sanjana et al. as well as the FLASH assembly process described by Reyon et al."

|
Scooped by
dromius
June 29, 2012 9:58 AM
|
DNA built from modular repeats presents a challenge for gene synthesis. We present a solid surface-based sequential ligation approach, which we refer to as iterative capped assembly (ICA), that adds DNA repeat monomers individually to a growing chain while using hairpin ‘capping’ oligonucleotides to block incompletely extended chains, greatly increasing the frequency of full-length final products. Applying ICA to a model problem, construction of custom transcription activator-like effector nucleases (TALENs) for genome engineering, we demonstrate efficient synthesis of TALE DNA-binding domains up to 21 monomers long and their ligation into a nuclease-carrying backbone vector all within 3 h. We used ICA to synthesize 20 TALENs of varying DNA target site length and tested their ability to stimulate gene editing by a donor oligonucleotide in human cells. All the TALENS show activity, with the ones >15 monomers long tending to work best. Since ICA builds full-length constructs from individual monomers rather than large exhaustive libraries of pre-fabricated oligomers, it will be trivial to incorporate future modified TALE monomers with improved or expanded function or to synthesize other types of repeat-modular DNA where the diversity of possible monomers makes exhaustive oligomer libraries impractical.

|
Scooped by
dromius
June 22, 2012 7:40 PM
|
Dr. Jon Chesnut, a Research Fellow in synthetic biology at Life Technologies, discusses his work on their new molecules, called TAL effectors...

|
Scooped by
dromius
May 22, 2012 5:01 AM
|
"This post on Biofortified asks for this breeding technology to be distinguished from transgenics – the technology that produces GMOs (or GM crops). Somehow I think this is wishful thinking because the genome has been modified and as the issue of GMO is more about values than the science, I suspect the same values that cause people to reject GMOs will be applied to the (TAL) effector technology. It will still be considered unnatural, and if it is as effective as predicted big companies will use it to their advantage. They will still questions its safety and potential effect on the environment….and so on"

|
Scooped by
dromius
May 17, 2012 5:19 AM
|
Instead of adding a sentence or two to the genome book, as is done by standard genetic modification (GM) approaches, they removed a few letters; the rice varieties they generated lack anywhere from 3 to 57 bases in their genomes. Thus, the rice plants generated by Li et al. do not
contain extraneous DNA and cannot by any reasonable definition be considered “GMOs.”

|
Scooped by
dromius
May 16, 2012 4:36 AM
|
The ability to engineer biological circuits that process and respond to complex cellular signals has the potential to impact many areas of biology and medicine. Transcriptional activator-like effectors (TALEs) have emerged as an attractive component for engineering these circuits, as TALEs can be designed de novo to target a given DNA sequence. Currently, however, the use of TALEs is limited by degeneracy in the site-specific manner by which they recognize DNA. Here, we propose an algorithm to computationally address this problem. We apply our algorithm to design 180 TALEs targeting 20 bp cognate binding sites that are at least 3 nt mismatches away from all 20 bp sequences in putative 2 kb human promoter regions. We generated eight of these synthetic TALE activators and showed that each is able to activate transcription from a targeted reporter. Importantly, we show that these proteins do not activate synthetic reporters containing mismatches similar to those present in the genome nor a set of endogenous genes predicted to be the most likely targets in vivo. Finally, we generated and characterized TALE repressors comprised of our orthogonal DNA binding domains and further combined them with shRNAs to accomplish near complete repression of target gene expression.

|
Scooped by
dromius
May 13, 2012 12:41 PM
|
...The described technique complies with mutagenesis in terms of § 3 No. 3b GenTG. According to these terms mutagenesis is not a procedure resulting in genetic modification. Thus, the ZKBS does not regard the resulting organisms as organisms modified by gene technology....

|
Scooped by
dromius
May 11, 2012 2:53 PM
|
A discussion that followed the landmark publication of Nature Biotechnology paper describing a disease resistant rice strain that was deleted for some DNA using TALEN technology http://tinyurl.com/cbs6gn6

|
Scooped by
dromius
May 8, 2012 12:41 PM
|
High-efficiency TALEN-based gene editing produces disease-resistant rice
http://www.nature.com/nbt/journal/v30/n5/full/nbt.2199.html?WT.ec_id=NBT-201205 from the Yang lab. Transcription activator–like (TAL) effectors of Xanthomonas oryzae pv. oryzae (Xoo) contribute to pathogen virulence by transcriptionally activating specific rice disease-susceptibility (S) genes. TAL effector nucleases (TALENs)—fusion proteins derived from the DNA recognition repeats of native or customized TAL effectors and the DNA cleavage domains of FokI—have been used to create site-specific gene modifications in plant cells, yeast, animals and even human pluripotent cells. Here, we exploit TALEN technology to edit a specific S gene in rice to thwart the virulence strategy of X. oryzae and thereby engineer heritable genome modifications for resistance to bacterial blight, a devastating disease in a crop that feeds half of the world's population.

|
Scooped by
dromius
April 30, 2012 3:03 AM
|
Genome engineering (GE), an emerging discipline in which a DNA sequence is altered at a single position, has a wide variety of potential uses, such as the correction of gene sequences in patients suffering from genetic diseases, the modification or insertion of genes in plants, and the generation of unique cell lines for treatment of diseases such as cancer. GE requires the development of molecular tools that can search out and bind to one unique site within a complex genome while avoiding 'off target' interactions across the remaining billions of DNA bases present in a cell’s nucleus.
|



 Your new post is loading...
Your new post is loading...