Your new post is loading...

|
Scooped by
?
February 26, 2018 10:21 PM
|
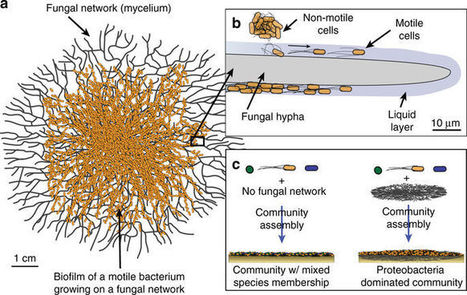
Most studies of bacterial motility have examined small-scale (micrometer–centimeter) cell dispersal in monocultures. However, bacteria live in multispecies communities, where interactions with other microbes may inhibit or facilitate dispersal. Here, we demonstrate that motile bacteria in cheese rind microbiomes use physical networks created by filamentous fungi for dispersal, and that these interactions can shape microbial community structure. Serratia proteamaculans and other motile cheese rind bacteria disperse on fungal networks by swimming in the liquid layers formed on fungal hyphae. RNA-sequencing, transposon mutagenesis, and comparative genomics identify potential genetic mechanisms, including flagella-mediated motility, that control bacterial dispersal on hyphae. By manipulating fungal networks in experimental communities, we demonstrate that fungal-mediated bacterial dispersal can shift cheese rind microbiome composition by promoting the growth of motile over non-motile community members. Our single-cell to whole-community systems approach highlights the interactive dynamics of bacterial motility in multispecies microbiomes.

|
Scooped by
?
February 26, 2018 12:56 PM
|
Anthocyanins are a class of brightly colored, glycosylated flavonoid pigments that imbue their flower and fruit host tissues with hues of predominantly red, orange, purple, and blue. Although all anthocyanins exhibit pH-responsive photochemical changes, distinct structural decorations on the core anthocyanin skeleton also cause dramatic color shifts, in addition to improved stabilities and unique pharmacological properties. In this work, we report for the first time the extension of the reconstituted plant anthocyanin pathway from (+)-catechin to O-methylated anthocyanins in a microbial production system, an effort which requires simultaneous co-option of the endogenous metabolites UDP-glucose and S-adenosyl-l-methionine (SAM or AdoMet). Anthocyanin O-methyltransferase (AOMT) orthologs from various plant sources were co-expressed in Escherichia coli with Petunia hybrida anthocyanidin synthase (PhANS) and Arabidopsis thaliana anthocyanidin 3-O-glucosyltransferase (At3GT). Vitis vinifera AOMT (VvAOMT1) and fragrant cyclamen ‘Kaori-no-mai’ AOMT (CkmOMT2) were found to be the most effective AOMTs for production of the 3′-O-methylated product peonidin 3-O-glucoside (P3G), attaining the highest titers at 2.4 and 2.7 mg/L, respectively. Following modulation of plasmid copy number and optimization of VvAOMT1 and CkmOMT2 expression conditions, production was further improved to 23 mg/L using VvAOMT1. Finally, CRISPRi was utilized to silence the transcriptional repressor MetJ in order to deregulate the methionine biosynthetic pathway and improve SAM availability for O-methylation of cyanidin 3-O-glucoside (C3G), the biosynthetic precursor to P3G. MetJ repression led to a final titer of 51 mg/L (56 mg/L upon scale-up to shake flask), representing a twofold improvement over the non-targeting CRISPRi control strain and 21-fold improvement overall. An E. coli strain was engineered for production of the specialty anthocyanin P3G using the abundant and comparatively inexpensive flavonol precursor, (+)-catechin. Furthermore, dCas9-mediated transcriptional repression of metJ alleviated a limiting SAM pool size, enhancing titers of the methylated anthocyanin product. While microbial production of P3G and other O-methylated anthocyanin pigments will likely be valuable to the food industry as natural food and beverage colorants, we expect that the strain constructed here will also prove useful to the ornamental plant industry as a platform for evaluating putative anthocyanin O-methyltransferases in pursuit of bespoke flower pigment compositions.

|
Scooped by
?
February 26, 2018 2:03 AM
|
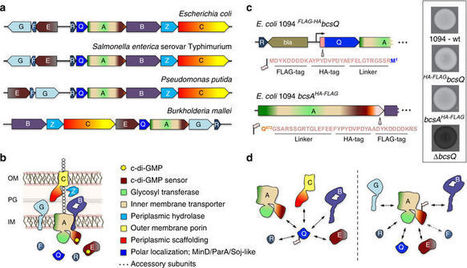
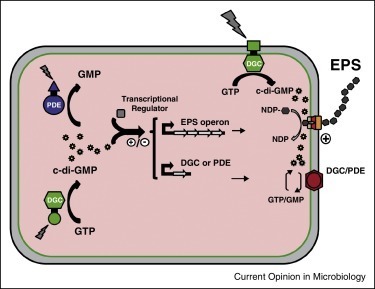
Secreted exopolysaccharides present important determinants for bacterial biofilm formation, survival, and virulence. Cellulose secretion typically requires the concerted action of a c-di-GMP-responsive inner membrane synthase (BcsA), an accessory membrane-anchored protein (BcsB), and several additional Bcs components. Although the BcsAB catalytic duo has been studied in great detail, its interplay with co-expressed subunits remains enigmatic. Here we show that E. coli Bcs proteins partake in a complex protein interaction network. Electron microscopy reveals a stable, megadalton-sized macromolecular assembly, which encompasses most of the inner membrane and cytosolic Bcs components and features a previously unobserved asymmetric architecture. Heterologous reconstitution and mutational analyses point toward a structure–function model, where accessory proteins regulate secretion by affecting both the assembly and stability of the system. Altogether, these results lay the foundation for more comprehensive models of synthase-dependent exopolysaccharide secretion in biofilms and add a sophisticated secretory nanomachine to the diverse bacterial arsenal for virulence and adaptation.

|
Scooped by
?
February 26, 2018 12:59 AM
|
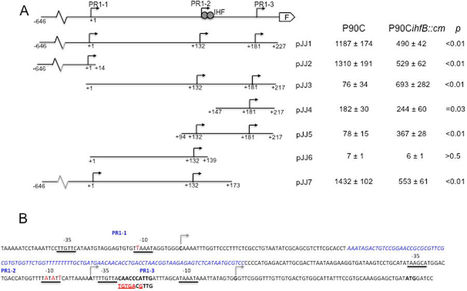
In this study we characterise three tandem promoters (PR1-1, PR1-2 and PR1-3) within the PR1 regulatory region of the Escherichia coli kps capsule gene cluster. Transcription from promoter PR1-2 was dependent on the activity of the upstream promoter PR1-1, which activated PR1-2 via transcription coupled DNA supercoiling. During growth at 37 °C a temporal pattern of transcription from all three promoters was observed with maximum transcriptional activity evident during mid-exponential phase followed by a sharp decrease in activity as the cells enter stationary phase. The growth phase dependent transcription was regulated by Integration Host Factor (IHF), which bound within the PR1 region to repress transcription from PR1-2 and PR1-3. This pattern of transcription was mirrored by growth phase dependent expression of the K1 capsule. Overall these data reveal a complex pattern of transcriptional regulation for an important virulence factor with IHF playing a role in regulating growth phase expression.

|
Scooped by
?
February 23, 2018 6:15 PM
|
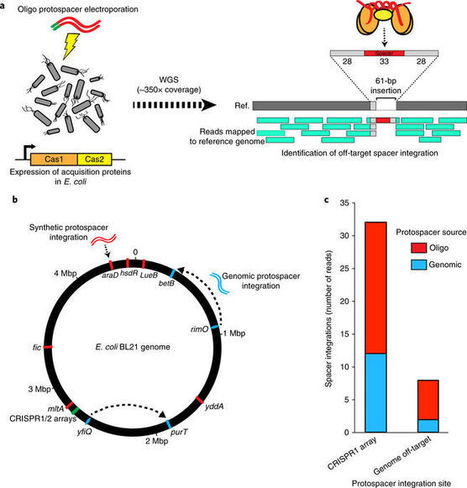
The adaptation phase of CRISPR–Cas immunity depends on the precise integration of short segments of foreign DNA (spacers) into a specific genomic location within the CRISPR locus by the Cas1–Cas2 integration complex. Although off-target spacer integration outside of canonical CRISPR arrays has been described in vitro, no evidence of non-specific integration activity has been found in vivo. Here, we show that non-canonical off-target integrations can occur within bacterial chromosomes at locations that resemble the native CRISPR locus by characterizing hundreds of off-target integration locations within Escherichia coli. Considering whether such promiscuous Cas1–Cas2 activity could have an evolutionary role through the genesis of neo-CRISPR loci, we combed existing CRISPR databases and available genomes for evidence of off-target integration activity. This search uncovered several putative instances of naturally occurring off-target spacer integration events within the genomes of Yersinia pestis and Sulfolobus islandicus. These results are important in understanding alternative routes to CRISPR array genesis and evolution, as well as in the use of spacer acquisition in technological applications.

|
Scooped by
?
February 23, 2018 5:33 PM
|
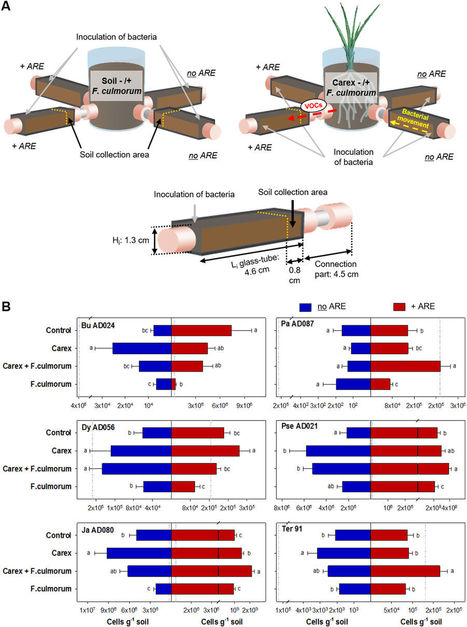
Plants release a wide set of secondary metabolites including volatile organic compounds (VOCs). Many of those compounds are considered to function as defense against herbivory, pests, and pathogens. However, little knowledge exists about the role of belowground plant VOCs for attracting beneficial soil microorganisms. We developed an olfactometer system to test the attraction of soil bacteria by VOCs emitted by Carex arenaria roots. Moreover, we tested whether infection of C. arenaria with the fungal pathogen Fusarium culmorum modifies the VOCs profile and bacterial attraction. The results revealed that migration of distant bacteria in soil towards roots can be stimulated by plant VOCs. Upon fungal infection, the blend of root VOCs changed and specific bacteria with antifungal properties were attracted. Tests with various pure VOCs indicated that those compounds can diffuse over long distance but with different diffusion abilities. Overall, this work highlights the importance of plant VOCs in belowground long-distance plant–microbe interactions.

|
Scooped by
?
February 23, 2018 5:10 PM
|
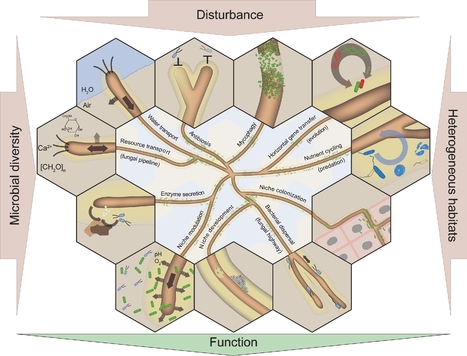
Fungi and bacteria are found living together in a wide variety of environments. Their interactions are significant drivers of many ecosystem functions and are important for the health of plants and animals. A large number of fungal and bacterial families are engaged in complex interactions that lead to critical behavioural shifts of the microorganisms ranging from mutualism to pathogenicity. The importance of bacterial-fungal interactions (BFI) in environmental science, medicine and biotechnology has led to the emergence of a dynamic and multidisciplinary research field that combines highly diverse approaches including molecular biology, genomics, geochemistry, chemical and microbial ecology, biophysics and ecological modelling. In this review, we discuss most recent advances that underscore the roles of BFI across relevant habitats and ecosystems. A particular focus is placed on the understanding of BFI within complex microbial communities and in regards of the metaorganism concept. We also discuss recent discoveries that clarify the (molecular) mechanisms involved in bacterial-fungal relationships, and the contribution of new technologies to decipher generic principles of BFI in terms of physical associations and molecular dialogues. Finally, we discuss future directions for researches in order to catalyse a synergy within the BFI research area and to resolve outstanding questions.

|
Scooped by
?
February 23, 2018 4:42 PM
|

|
Scooped by
?
February 23, 2018 12:44 PM
|

|
Scooped by
?
February 23, 2018 11:19 AM
|
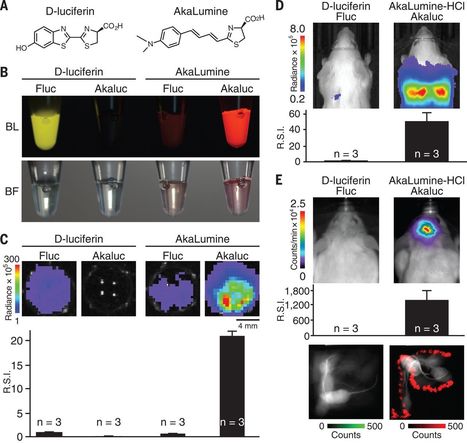
Bioluminescence imaging is a tremendous asset to medical research, providing a way to monitor living cells noninvasively within their natural environments. Advances in imaging methods allow researchers to measure tumor growth, visualize developmental processes, and track cell-cell interactions. Yet technical limitations exist, and it is difficult to image deep tissues or detect low cell numbers in vivo. Iwano et al. designed a bioluminescence imaging system that produces brighter emission by up to a factor of 1000 compared with conventional technology (see the Perspective by Nasu and Campbell). Individual tumor cells were successfully visualized in the lungs of mice. Small numbers of striatal neurons were detected in the brains of naturally behaving marmosets. The ability of the substrate to cross the blood-brain barrier should provide important opportunities for neuroscience research.
Science , this issue p. [935][1]; see also p. [868][2]
[1]: /lookup/doi/10.1126/science.aaq1067
[2]: /lookup/volpage/359/868?iss=6378
|

|
Scooped by
?
February 26, 2018 5:50 PM
|
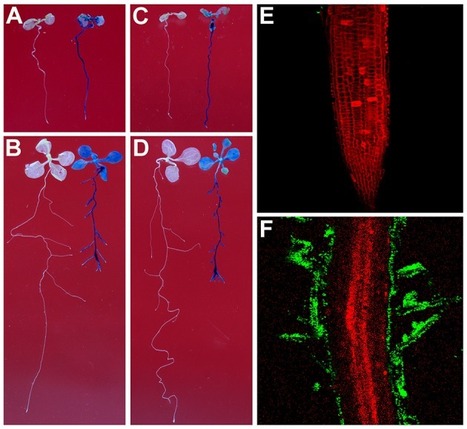
Rhizobium sp. IRBG74 not only nodulates Sesbania cannabina but also can enhance rice growth; however, the underlying molecular mechanisms are not clear. Here, we show that Rhizobium sp. IRBG74 colonizes the roots of Arabidopsis thaliana, which leads to inhibition in the growth of main root but enhancement in the formation of lateral roots. The promotion of lateral root formation by Rhizobium sp. IRBG74 in the fls2-1 mutant, which is insensitive to flagellin, is similar to the wild-type plant, while the auxin response deficient mutant tir1-1 is significantly less sensitive to Rhizobium sp. IRBG74 than the wild type in terms of the inhibition of main root elongation and the promotion of lateral root formation. Further transcriptome analysis of Arabidopsis roots inoculated with Rhizobium sp. IRBG74 revealed differential expression of 50 and 211 genes at 24 and 48 h, respectively, and a majority of these genes are involved in auxin signaling. Consistent with the transcriptome analysis results, Rhizobium sp. IRBG74 treatment induces expression of the auxin responsive reporter DR5:GUS in roots. Our results suggest that in Arabidopsis Rhizobium sp. IRBG74 colonizes roots and promotes the lateral root formation likely through modulating auxin signaling. Our work provides insight into the molecular mechanisms of interactions between legume-nodulating rhizobia and non-legume plants.

|
Scooped by
?
February 26, 2018 10:25 AM
|
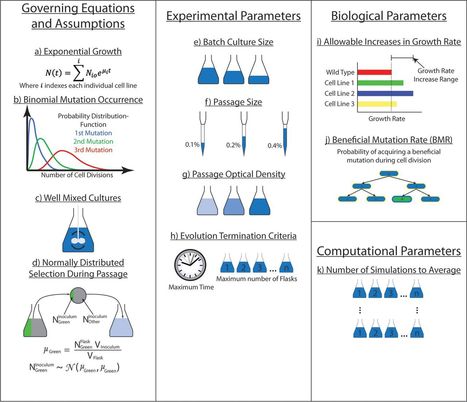
The occurrence of mutations is a cornerstone of the evolutionary theory of adaptation, capitalizing on the rare chance that a mutation confers a fitness benefit. Natural selection is increasingly being leveraged in laboratory settings for industrial and basic science applications. Despite increasing deployment, there are no standardized procedures available for designing and performing adaptive laboratory evolution (ALE) experiments. Thus, there is a need to optimize the experimental design, specifically for determining when to consider an experiment complete and for balancing outcomes with available resources (i.e., laboratory supplies, personnel, and time). To design and to better understand ALE experiments, a simulator, ALEsim, was developed, validated, and applied to the optimization of ALE experiments. The effects of various passage sizes were experimentally determined and subsequently evaluated with ALEsim, to explain differences in experimental outcomes. Furthermore, a beneficial mutation rate of 10−6.9 to 10−8.4 mutations per cell division was derived. A retrospective analysis of ALE experiments revealed that passage sizes typically employed in serial passage batch culture ALE experiments led to inefficient production and fixation of beneficial mutations. ALEsim and the results described here will aid in the design of ALE experiments to fit the exact needs of a project while taking into account the resources required and will lower the barriers to entry for this experimental technique.

|
Scooped by
?
February 26, 2018 1:49 AM
|
Quorum sensing (QS) is a cell–cell communication process that enables bacteria to track cell population density and orchestrate collective behaviors. QS relies on the production and detection of, and the response to, extracellular signal molecules called autoinducers. In Vibrio cholerae, multiple QS circuits control pathogenesis and biofilm formation. Here, we identify and characterize a new QS autoinducer–receptor pair. The autoinducer is 3,5-dimethylpyrazin-2-ol (DPO). DPO is made from threonine and alanine, and its synthesis depends on threonine dehydrogenase (Tdh). DPO binds to and activates a transcription factor, VqmA. The VqmA–DPO complex activates expression of vqmR, which encodes a small regulatory RNA. VqmR represses genes required for biofilm formation and toxin production. We propose that DPO allows V. cholerae to regulate collective behaviors to, among other possible roles, diversify its QS output during colonization of the human host.

|
Scooped by
?
February 25, 2018 12:34 PM
|
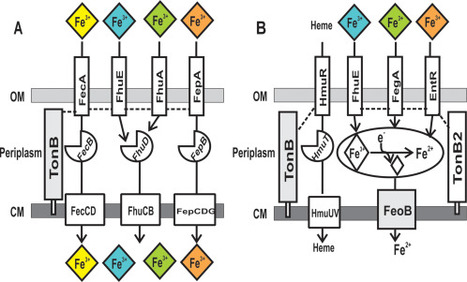
Under iron limitation, bacteria scavenge ferric (Fe3+) iron bound to siderophores or other chelates from the environment to fulfill their nutritional requirement. In gram-negative bacteria, the siderophore uptake system prototype consists of an outer membrane transporter, a periplasmic binding protein and a cytoplasmic membrane transporter, each specific for a single ferric siderophore or siderophore family. Here, we show that spontaneous single gain-of-function missense mutations in outer membrane transporter genes of Bradyrhizobium japonicum were sufficient to confer on cells the ability to use synthetic or natural iron siderophores, suggesting that selectivity is limited primarily to the outer membrane and can be readily modified. Moreover, growth on natural or synthetic chelators required the cytoplasmic membrane ferrous (Fe2+) iron transporter FeoB, suggesting that iron is both dissociated from the chelate and reduced to the ferrous form within the periplasm prior to cytoplasmic entry. The data suggest rapid adaptation to environmental iron by facile mutation of selective outer membrane transporter genes and by non-selective uptake components that do not require mutation to accommodate new iron sources.

|
Scooped by
?
February 23, 2018 6:12 PM
|
Actinobacteria is a phylum of gram-positive bacteria, and members of bacteria belonging to this phylum are classified into six classes, namely Acidimicrobiia, Actinobacteria, Coriobacteriia, Nitriliruptoria, Rubrobacteria, and Thermoleophilia. Among the six different classes, members of the actinobacteria class are the most dominant and contain one of the largest genera, Streptomyces, with 961 distinct species. Members of phylum actinobacteria are ubiquitous in nature and have been isolated from various extreme environments (high temperatures, pH, salinities, pressure, and drought) and are associated with plants growing in different habitats. The rhizospheric actinobacteria are the most dominant in nature and they are of great economic importance to humans due to their huge contributions to soil systems. Based on a comprehensive literature analysis members of phylum actinobacteria have been reported from different genera such as Acidimicrobium, Actinomyces, Arthrobacter, Bifidobacterium, Cellulomonas, Clavibacter, Corynebacterium, Frankia, Microbacterium, Micrococcus, Mycobacterium, Nocardia, Propionibacterium, Pseudonocardia, Rhodococcus, Sanguibacter, and Streptomyces. Actinobacteria can be utilized as bioinoculants for sustainable agriculture as they can enhance plant growth and yield both by directly promoting plant growth by the fixation of atmospheric nitrogen, the solubilization of minerals such as phosphorus, potassium, and zinc, the production of siderophores and plant growth hormones such cytokinin, auxin, and gibberellins, and by indirect plant growth promotion via production of antagonistic substances by inducing resistance against plant pathogens. The actinobacteria has been proved to have immense importance in biotechnological applications in various industrial and agricultural processes that are discussed in this chapter.

|
Scooped by
?
February 23, 2018 5:28 PM
|
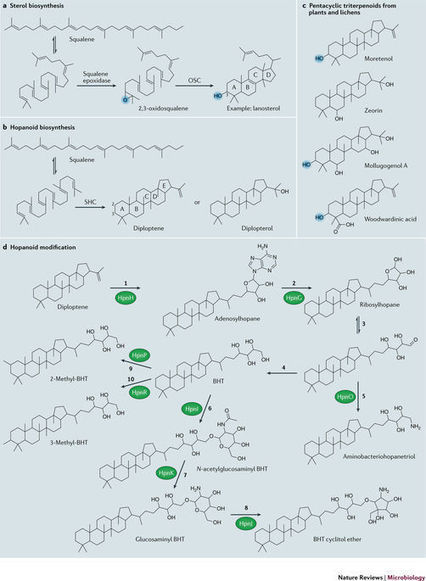
Hopanoids, which resemble sterols and are found in the membranes of diverse bacteria, have left an extensive molecular fossil record. Today, hopanoid-producing bacteria remain abundant in various ecosystems, such as the rhizosphere. In vitro, hopanoids contribute to bacterial stress resistance, which may help explain their ability to facilitate beneficial plant–bacteria interactions. However, given that hopanoids can also serve as carriers for plant hormones and that plants themselves make hopanoid-like compounds, it is likely that other mechanisms are additionally at play. In closing, we remind the reader of the words of John F. Kennedy when explaining what was needed to send astronauts to the Moon. “We choose to go to the Moon! ... We choose to go to the Moon in this decade and do the other things, not because they are easy, but because they are hard; because that goal will serve to organize and measure the best of our energies and skills...”. Replacing the words 'go to the Moon' with 'study lipids' provides fitting inspiration for tackling one of the next great challenges in microbiology. It is time for lipids to get the attention they deserve, and the study of hopanoids is an excellent place to begin.

|
Scooped by
?
February 23, 2018 4:53 PM
|
Four nucleotide bases carry all the information an organism needs to live and function. Can this limited set of bases be expanded to carry more information? Now, a new study by Floyd Romesberg at The Scripps Research Institute reports that introducing unnatural nucleotides into DNA results in the incorporation of novel amino acids into proteins in E.coli. While other research groups have engineered cells to incorporate unnatural amino acids into cellular proteins, Romesberg focused on expanding the information stored in DNAso that semi-synthetic organisms could create new proteins and perform unnatural functions. “The natural system was there, and I wanted to augment the natural system,” said Romesberg.

|
Scooped by
?
February 23, 2018 3:53 PM
|
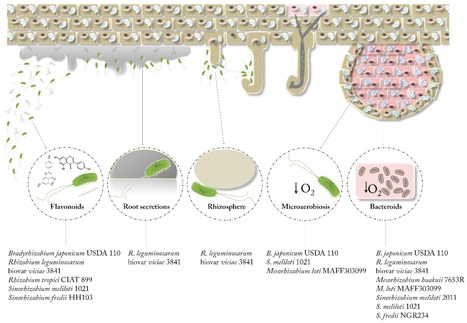
Simultaneous quantification of transcripts of the whole bacterial genome allows the analysis of the global transcriptional response under changing conditions. RNA-seq and microarrays are the most used techniques to measure these transcriptomic changes, and both complement each other in transcriptome profiling. In this review, we exhaustively compiled the symbiosis-related transcriptomic reports (microarrays and RNA sequencing) carried out hitherto in rhizobia. This review is specially focused on transcriptomic changes that takes place when five rhizobial species, Bradyrhizobium japonicum (=diazoefficiens) USDA 110, Rhizobium leguminosarum biovar viciae 3841, Rhizobium tropici CIAT 899, Sinorhizobium (=Ensifer) meliloti 1021 and S. fredii HH103, recognize inducing flavonoids, plant-exuded phenolic compounds that activate the biosynthesis and export of Nod factors (NF) in all analysed rhizobia. Interestingly, our global transcriptomic comparison also indicates that each rhizobial species possesses its own arsenal of molecular weapons accompanying the set of NF in order to establish a successful interaction with host legumes.

|
Scooped by
?
February 23, 2018 11:23 AM
|
|

 Your new post is loading...
Your new post is loading...