Your new post is loading...

|
Scooped by
mhryu@live.com
March 7, 3:59 PM
|
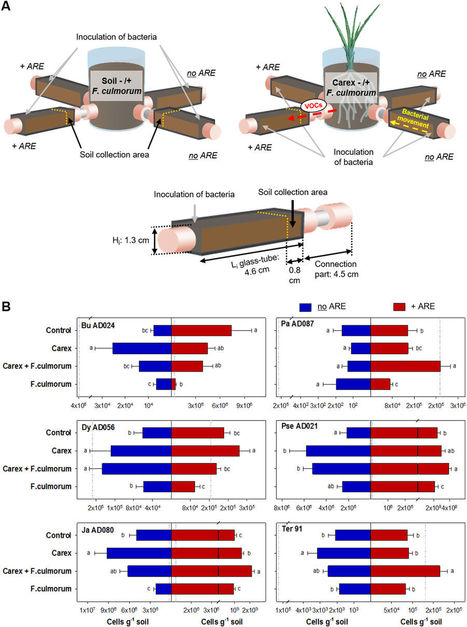
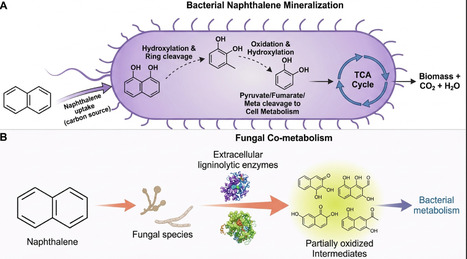
Naphthalene, a widely detected polycyclic aromatic hydrocarbon (PAH), is among the 16 priority PAHs identified as major environmental hazards due to its persistence, ubiquity, and toxicity to ecosystems and human health. Its occurrence in crude oil, combustion residues, vehicle emissions, and household products highlights the urgent need for sustainable remediation strategies. Microbial-based bioremediation stands out as an eco-friendly and cost-effective approach that harnesses the metabolic versatility of diverse microorganisms, their genes, and enzymes responsible for naphthalene degradation. Recent advances in omics technologies and high-throughput sequencing have expanded our understanding of novel microbial taxa, metabolic pathways, and stress responses under naphthalene exposure. Complementarily, computational modelling, in silico tools, machine learning, and systems biology have enabled the prediction of degradation dynamics and the design of synthetic microbial consortia optimised for field use. Despite these advances, challenges such as environmental fluctuations, co-contaminant effects, and the gap between laboratory and field outcomes remain. Overcoming these requires an integrative framework that connects microbial ecology, omics insights, and computational modelling. This review consolidates current knowledge on microbial degradation of naphthalene, emphasising key taxa, genes, and pathways, and highlights how omics, in silico tools and systems biology can drive sustainable remediation in the Anthropocene.

|
Scooped by
mhryu@live.com
March 7, 3:48 PM
|
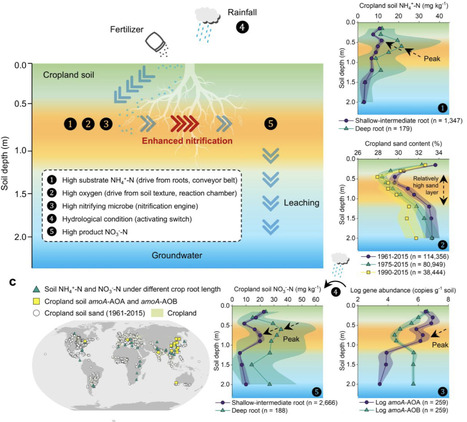
The excessive application of nitrogen fertilizers in croplands drives nitrification-induced nitrogen losses, accelerating soil degradation and groundwater nitrate pollution. By synthesizing global nitrogen profiles and field studies, we identify a previously overlooked “enhanced nitrification layer (ENL)” at approximately 0.6 m depth (ranging from 0.3 to 0.9 m) across global croplands. The formation of the ENL relies on the interplay of four key processes: root-mediated nitrogen transport (the conveyor belt), an oxygenated microenvironment sustained by soil texture (the reaction chamber), nitrifier communities (the nitrification engine), and favorable hydrological conditions (the activating switch). This ENL acts as a biogeochemical hotspot, driving subsoil pH declines and nitrate leaching to groundwater. Its identification establishes a precise subsurface target, expanding nitrogen management to address the critical depth of peak transformation alongside surface measures, thereby advancing agroecosystem sustainability. Global cropland subsoil hides a “biogeochemical engine” where crop roots, soil texture, nitrifier communities, and hydrological conditions interact to fuel nitrification, silently driving soil acidification and groundwater nitrate pollution.

|
Scooped by
mhryu@live.com
March 7, 3:33 PM
|
Protein binders are fundamental tools in chemical biology, key components of biotechnologies, and the foundation of biologics-based medicines. However, no binder discovery method has achieved the fidelity, speed, and cost-effectiveness required for routine laboratory use. We argue that the field stands at an inflection point. In vitro display platforms are now mature but have continued evolving through macrocyclization methods, covalent chemistries, and massive quantitative datasets—expanding chemical space while deepening mechanistic understanding of selection outcomes. Computational de novo design—which has exploded in power over the past few years—is transitioning from methods requiring specialized expertise to more accessible platforms. Finally, in vivo directed evolution technologies are emerging as a critical frontier, which may deliver the throughput and fidelity needed to finally democratize binder discovery. This review summarizes these advances and points toward a future where binder generation is fast, accessible, and a key driver of the next phase of biological research.

|
Scooped by
mhryu@live.com
March 7, 3:17 PM
|
Conventional chemical pesticides have long protected global crop yields, but their environmental and health impacts, including contamination, biodiversity loss, residue accumulation, and pest resistance, demand sustainable alternatives. RNA interference (RNAi) represents a genetic breakthrough in crop protection, enabling precise, sequence-specific silencing of essential pest genes via double-stranded RNA (dsRNA). Innovations such as nanoparticle-based delivery, host-induced gene silencing (HIGS), and spray-induced gene silencing (SIGS) have transformed RNAi into a practical, field-ready technology. Integration with multi-omics and computational biology accelerates target discovery and enhances specificity and scalability. Beyond pest management, RNAi applications extend to vector control and pollinator protection, exemplifying One Health principles. RNAi-based pesticides therefore, redefine sustainable agriculture through molecular precision and cross-sectoral genetic innovation.

|
Scooped by
mhryu@live.com
March 7, 3:03 PM
|
Protein is the main structural and functional component of cells, making it crucial for the survival of all living organisms. Yet mammalian herbivores and omnivores often consume diets deficient in the amount of protein required for growth, homeostasis, and reproduction. To compensate, mammals likely rely on their gut microbiota to synthesize essential amino acids (AAESS), particularly during periods of dietary protein limitation. We quantified the contribution of microbially synthesized AAESS to skeletal muscle in captive, wild-derived deer mice (Peromyscus maniculatus) fed diets varying in macromolecular quantity and quality. Using amino acid carbon isotope (δ13C) analysis combined with genetic sequencing, we assessed the origin of AAESS incorporated into host muscle and identified gut microbial taxa with the genetic potential for AAESS biosynthesis. We estimate that up to 25% of host muscle AAESS were microbially derived, with greater microbial contributions in mice fed diets containing low protein or more complex macronutrients. Gut microbial populations with the genetic potential for AAESS biosynthesis were more abundant in mice with larger contributions of microbially-derived AAESS in their tissues. These results demonstrate the crucial and likely pervasive role the gut microbiome plays in host protein metabolism, especially in mammals facing seasonal or persistent dietary protein limitation.

|
Scooped by
mhryu@live.com
March 7, 12:51 PM
|
For much of the 20th century, enzyme kinetic analysis relied on deriving simplified rate equations under the steady-state approximation and later by analytical integration of differential equations for transient kinetics. This approach has since been surpassed by computational methods using numerical integration of rate equations to directly fit experimental data based on a complete user-defined model. This paradigm shift removes the constraints imposed by solving analytical equations, enabling far greater flexibility in experimental design and model complexity. Modern global fitting methods allow data from diverse experiments to be analyzed simultaneously using the minimum number of parameters supported by the information content of the data set. Global data fitting is more than just an algorithm for data analysis─it represents a fundamental change in how we design and interpret experiments, and eliminates many of the restrictions, approximations, and ambiguities inherent to equation-based analyses. In this review, we describe the principles and practice of global data fitting, compare the outcomes to conventional equation-based methods, and demonstrate its power through examples involving multiple experiments with distinct conditions and readouts. We explain why the common practice of making measurements in triplicate introduces uncertainty and we outline advanced methods for rigorously estimating errors in measurement and for establishing robust confidence limits on fitted parameters.

|
Scooped by
mhryu@live.com
March 7, 12:36 PM
|
Intermicrobial and host–microbial interactions are critical for the functioning of the gut microbiome, but few tools are available to measure these interactions in situ. Here we report a method for broad spatial sampling of microbiome–host interactions in the gut at high resolution (1 µm). This method combines enzymatic in situ polyadenylation of both bacterial and host RNA with spatial RNA sequencing to increase bacterial RNA recovery and enable transcriptomic analysis of low-abundance and spatially restricted microbial taxa. We benchmark the method against existing spatial transcriptomic workflows, demonstrating improved sensitivity and resolution. Application of this method in a mouse model of intestinal neoplasia revealed the biogeography of the mouse gut microbiome as function of location in the intestine, frequent strong intermicrobial interactions at short length scales and tumor-associated changes in the architecture of the host–microbiome interface. This method is compatible with widely available commercial platforms for spatial RNA sequencing and can therefore be readily adopted to study the role of short-range, bidirectional host–microbe interactions in microbiome health and disease. Bulk polyadenylation improves the capture of microbial RNA, enabling host–microorganism interactions to be visualized at a high resolution when combined with commercially available spatial transcriptomics platforms.

|
Scooped by
mhryu@live.com
March 7, 12:23 PM
|
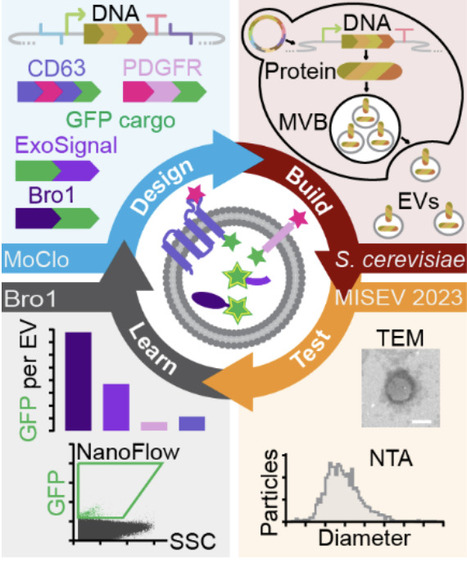
Extracellular vesicles (EVs) hold great promise as therapeutic delivery vehicles, leveraging their natural role as mediators of intercellular communication in all organisms studied. However, many barriers must be overcome to realize their full potential. Saccharomyces cerevisiae is an attractive chassis organism to explore solutions: It is used for drug biomanufacturing, it is amenable to complex genetic engineering, and their EVs can drive responses in human cells. To further develop this prospect, we sought to genetically modify S. cerevisiae EVs by devising a research framework amenable to iterative design, build, test, learn cycles, a core principle of synthetic biology. Using this approach, we focused on identifying new scaffolds, proteins that load cargoes into EVs, from a small pool of candidates. We first optimized a modular cloning strategy, called EVclo, for plasmid and genome-integrated candidate gene expression. Candidate genes were fused to EGFP, and after confirming expression in cells, we showed that scaffold-EFGP proteins colocalized with mRuby2-tagged Nhx1, a biomarker of multivesicular bodies, presumed sites of EV biogenesis. We triggered release of EVs by heat stress, isolated these EVs by ultrafiltration and size exclusion chromatography, and confirmed the presence of exosome-sized EVs in all samples. We find that candidate scaffold proteins did not affect EV size, morphology or titers. Further analysis of these samples indicated that some EGFP-tagged scaffolds are present in EVs: Bro1, a yeast ortholog of ALIX, was most abundant and ExoSignal showed highest enrichment of the human candidates. In all, we conclude that Bro1 is a good scaffold for future engineering strategies, and that human proteins can be sorted into yeast EVs suggesting conservation of the sorting machinery and demonstrating that yeast EVs can be humanized. This synthetic biology-based, proof-of-concept study establishes S. cerevisiae as a platform to engineer and bioproduce designer EVs for many applications.

|
Scooped by
mhryu@live.com
March 7, 12:16 PM
|
Microbial single-amplified genome (SAG) sequencing technologies have elevated microbial research resolution to the single-cell level. However, neither upstream data processing nor downstream analysis has been fully developed, greatly limiting the research in strain level. Herein, we developed MetaSAG (Multi-level Exploration and Taxonomic Analysis of microbial Single-Amplified Genomes), which enables accurate and rapid taxonomic classification of microbial SAGs. MetaSAG outperforms existing method in species classification certainty, computational efficiency, and sensitivity of low abundance species identification. In addition, MetaSAG enables species-level functional analysis, as well as strain-level evolutionary analysis. With the help of MetaSAG, we discovered the parasitic relationship between phages and bacteria, identifying multiple susceptible bacteria and a broad spectrum of phages. Furthermore, we developed MetaK-Lytic (k-mer-based meta-learning framework to predict phage lytic ability) to achieve accurate prediction of phage lytic activity based on 31-mer short sequences, which is well adapted to the characteristics of incomplete SAG sequences. Overall, we offer a comprehensive integrated tool that can parse microbial SAG data from raw data to the strain level to decipher the functional ecology of microbial dark matter, with broad implications for microbial ecology and phage therapy (https://github.com/liangcheng-hrbmu/MetaSAG).

|
Scooped by
mhryu@live.com
March 7, 12:00 PM
|
Proteins are built from 20 canonical amino acids. It is interesting to explore whether proteins can be formed from significantly reduced amino acid alphabets. Our bioinformatics survey of UniProt (more than 250 M sequences) revealed that proteins composed of reduced amino acid alphabets (<~10) are extremely rare among existing proteins. Next, we used computational protein design to design proteins composed of all 1,013 possible alphabets of 2-10 early amino acids (Ala, Asp, Glu, Gly, Ile, Leu, Pro, Ser, Thr, and Val). The length of all proteins was 100 amino acid residues. Small amino acid alphabets preferred simple helices or helix bundles. Larger amino acid alphabets allowed for the design of more complex structures. A protein composed of 8 amino acids (Ala, Asp, Gly, Leu, Val, Ser, Thr, and Pro) was successfully experimentally verified. It belongs to fibronectin type III domain beta-sheet-rich architecture. Attempts to experimentally verify designs composed of 6 and 4 amino acids were unsuccessful. We show by a computational experiment without an experimental validation that inverse folding programs, namely ProteinMPNN, can stabilize designed proteins within the same amino acid alphabet. Our results show that globular proteins may have formed early in evolution. Furthermore, we show that it is possible to design proteins with interesting properties for biotechnology and synthetic biology.

|
Scooped by
mhryu@live.com
March 7, 11:53 AM
|
Drought reshapes plant root microbiota, yet the mechanistic drivers and consequences of this observation remain unclear. We discovered that suppression of host immunity and iron homeostasis is required for Streptomyces enrichment in roots during drought across diverse soils. Genetic and physiological manipulation of these host pathways confirmed their requirement in modulating Streptomyces root enrichment. Drought-induced suppression of iron uptake was conserved across the ~160 My monocot-eudicot divergence. Some Streptomyces strains enhanced plant growth and rescued iron uptake under drought. These benefits were uncoupled from Streptomyces root enrichment. They were instead shaped by intra-Streptomyces antagonism. We propose a two-step model: drought-driven down regulation of host iron and immune pathways enriches Streptomyces, while intra-genus dynamics fine-tune strain-level assembly and functional outcomes. Our data refine the idea that Streptomyces are enriched in roots during drought in response to a plant cry for help and consequently contribute to alleviation of this abiotic stress.

|
Scooped by
mhryu@live.com
March 7, 11:42 AM
|
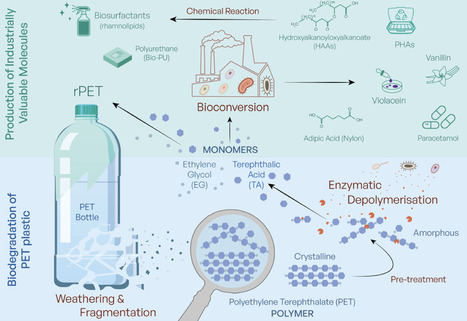
Plastics drive twin crises: persistent pollution and greenhouse gas emissions. Bio-based approaches using enzymes and microorganisms to depolymerise plastics and valorise monomers show promise but raise societal, ethical and regulatory questions central to Responsible Research and Innovation (RRI). In this Perspective, we reflect on RRI implications of bio-based plastic degradation, informed by stakeholder discussions across the plastics value chain and public engagement. We identify broad support alongside concerns about scalability, interaction with existing recycling, governance and containment of genetically modified organisms, management of additives and contaminants, and the roles of regulation and economic incentives in enabling adoption. Bio-based approaches to plastic degradation have been an intense area of research in recent years, and although they show great promise, they also raise societal, ethical and regulatory questions. Here the authors reflect on the Responsible Research and Innovation (RRI) implications of this growing field, sharing insights they have gained through engagement with stakeholders and the broader public.

|
Scooped by
mhryu@live.com
March 6, 6:11 PM
|
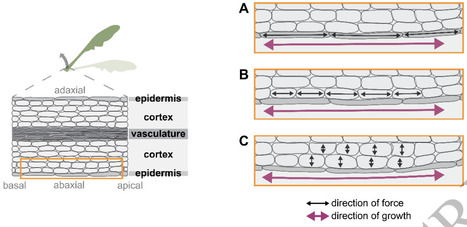
Plants use light both as a resource for photosynthesis and as a signal about their environment. In response to light cues, plants can move their organs via directional growth driven by cell expansion. In dense vegetation where light is available in spatially heterogeneous patterns, plants need to navigate this space to improve the position of their photosynthetic tissues. In canopies blue light irradiance and red to far-red light ratio decrease due to absorption by chloroplasts, and these changes regulate distinct processes within the plant. Changes in light environment are detected by cryptochrome and phytochrome photoreceptors, both regulating Phytochrome Interacting Factors (PIFs) and thereby enhancing elongation in hypocotyls, stems and leaves, and inducing upward leaf movement (hyponasty). An additional class of photoreceptors, phototropins, decode horizontal light gradients to produce directional growth towards the light source (phototropism). Here we review the current state of knowledge on these differential growth responses to light cues, with specific emphasis on the regulatory pathways that translate light signaling in differential cell expansion. Downstream of the photoreceptors, the phytohormone auxin induces cell growth in shoot tissues, but also other phytohormones contribute to balancing light responses. Cell expansion is regulated primarily at the level of cell walls and a comparison of different transcriptome datasets reveals that only a small group of cell wall modifying genes are tightly regulated by shade cues. It remains poorly understood which cell layers are causal to the initiation of cellular expansion. Here we combine insights from different differential growth behaviors in different species and organs to generate different hypotheses for the cellular underpinnings of light-driven leaf movements.
|

|
Scooped by
mhryu@live.com
March 7, 3:56 PM
|
Pollution causes disease and premature death globally. Various strategies, including biological methods like enzymes, are used to treat pollutants. The production of recombinant proteins in E. coli often results in their aggregation as inclusion bodies (IBs), enriched with the protein of interest. We developed a novel strategy for producing an immobilized bacterial laccase through self-assembly into catalytically active IBs (CatIBs) in E. coli. Our target was the outer spore-coat protein CotA from Bacillus clausii, a halotolerant and pH-stable laccase suitable for scalable applications in wastewater treatment. Two constructs were designed: Lac-17 (without fusion tags) and Lac-40EL (fused to a synthetic 35-amino-acid peptide, named as 40EL), both expressed in E. coli BL21(DE3). Both strains showed similar growth patterns, though Lac-17 yielded more total recombinant protein (~ 163 mg/L vs. ~127 mg/L). However, Lac-40EL formed CatIBs with up to 22-fold higher enzymatic activity than Lac-17, despite ~ 85% of the recombinant protein being in the insoluble fraction in both cases. Structural models suggest that the 40EL peptide forms an exposed double α-helix that promotes ordered aggregation without hindering the active site of the enzyme. Unlike the soluble fractions, the CatIBs retained their activity after three months at 4 °C. In azo dye decolorization assays with Eriochrome Black T and Congo Red, Lac-40EL CatIBs outperformed Lac-17, particularly in the presence of redox mediators. This work shows that rational peptide design can improve enzyme immobilization, activity, and stability through CatIBs formation. The resulting CatIBs self-assembled efficiently, were easily recovered, and demonstrated operational stability, supporting their potential for scalable production and novel applications in textile wastewater treatment.

|
Scooped by
mhryu@live.com
March 7, 3:41 PM
|
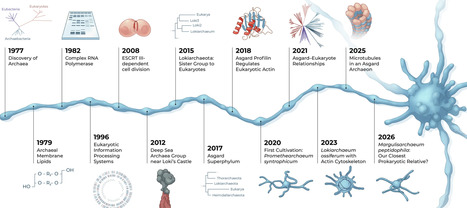
The discovery of Asgard archaea about a decade ago has greatly reshaped our understanding of archaeal evolution and the origin of eukaryotes. Asgards are currently thought to be the closest prokaryotic relatives of eukaryotes and to represent the archaeal host lineage that participated in the endosymbiotic event leading to the first eukaryotic cell. The presence of numerous eukaryotic signature proteins in Asgard genomes supports this view and provides important insights into the deep evolutionary roots of eukaryotic cellular complexity. However, the close relationship between archaea and eukaryotes had been observed for decades, based on features that are shared in different molecular processes. This review discusses the discovery of Asgard archaea in the broader context of archaeal molecular and cellular biology and highlights how earlier findings foreshadowed their emergence. Primarily targeted at newcomers to the field, the review provides an overview of evolutionary innovations across the Archaea domain and discusses molecular and cellular features of cultivated Asgard strains in light of previous archaeal research.

|
Scooped by
mhryu@live.com
March 7, 3:22 PM
|
Microbial synthesis provides a sustainable and efficient platform for pharmaceutical production, primarily through heterologous enzyme expression in engineered cell factories. However, this approach often suffers from intermediate diffusion, metabolic flux competition, and cytotoxic accumulation. Inspired by naturally evolved pathways where enzymes are spatially organized to enhance catalytic specificity and efficiency, artificial spatial assembly tools have been developed to overcome these limitations. This review comprehensively summarizes recent advances in such systems — including rational peptide linkers, peptide–protein interaction domains, nucleic acid scaffolds, and self-assembling elements — applied to diverse pharmaceuticals like antitumor drugs, antimalarials, and nutraceuticals. We further discuss key challenges and future directions to advance efficient and sustainable biosynthesis via spatial engineering.

|
Scooped by
mhryu@live.com
March 7, 3:15 PM
|
Microbial-induced calcite precipitation (MICP) offers a sustainable strategy for extending the service life of concrete through autonomous crack healing, yet the high alkalinity of cementitious environments restricts microbial viability. In this study, more than 200 indigenous bacterial isolates collected from extreme environments across Iran were systematically screened for urease and carbonic anhydrase (CA) activities. A dual-enzyme activity index (EAI) was developed to quantitatively rank their calcification potential. Four robust spore-forming strains—Bacillus subtilis, Sporosarcina pasteurii, Bacillus sphaericus, and the environmental isolate E10.2—were identified as top candidates based on high EAI values, sporulation capacity, and survival at pH 13.5. These strains retained at least 70% of their enzymatic activity after alkaline exposure and precipitated up to 89% more CaCO3 than controls. When incorporated into mortar, bio-treated specimens reached strength levels slightly exceeding the uncracked control under the tested conditions (46.8 MPa at 28 days compared to 34.2 MPa in cracked controls). Ultrasonic pulse velocity, SEM, and XRD analyses confirmed dense CaCO3 bridging within healed cracks. This study establishes a performance-based framework for selecting dual-enzyme-producing alkaliphilic bacteria for durable, self-healing concrete.

|
Scooped by
mhryu@live.com
March 7, 12:55 PM
|
Although it is increasingly recognized that anthropogenic chemicals modulate horizontal gene transfer (HGT), the nature of these interactions is often more complex than a simple promotion or inhibition. The potential for a single chemical to exert opposing, concentration-dependent effects represent a critical and less-explored frontier in microbial ecology. Here, we investigate the last-resort antibiotic polymyxin B, a membrane-targeting peptide, and reveal a concentration-dependent, biphasic regulation of plasmid conjugation. Sub-inhibitory concentrations (0.125-0.5 mg/L) consistently inhibited the transfer of antibiotic resistance genes (ARGs) by up to 65.4%, whereas bactericidal concentrations (≥ 1 mg/L) strongly promoted it by up to 15.9-fold. This regulatory switch is driven by distinct physiological states: low-level exposure triggers defensive responses including reduced membrane permeability, whereas high-level exposure causes catastrophic membrane damage, inducing a synergistic stress response involving oxidative damage (>2-fold ROS increase) and a surge in cellular energy (up to 83.0% ATP increase) that facilitates HGT. High-concentration polymyxin B also promotes plasmid transfer in complex microbial communities derived from activated-sludge biofilms. Our findings reveal a new paradigm for the interaction between chemical stressors and microbial evolution, demonstrating that the ecological impact of contaminants on HGT cannot be predicted by monotonic models and highlighting the role of environmental hotspots in shaping the dissemination of antibiotic resistome.

|
Scooped by
mhryu@live.com
March 7, 12:47 PM
|
Terpenoids are highly abundant and valuable natural products, yet high-throughput screening platforms for microbial producers remain scarce. To address this, we developed a microfluidic screening platform leveraging the intrinsic fluorescence of β-carotene. Using an engineered yeast chassis (YsL4) with a reinforced mevalonate pathway, we optimized cultivation conditions and achieved perfect (100%) phenotypic specificity in a 1:1 coencapsulation assay between producer (YsL4) and nonproducer (Ys011) strains. Screening a genome-wide CRISPRa/i library via multiround fluorescence-activated droplet sorting identified 15 target genes. Validation confirmed that 10 targets (5 for overexpression, 10 for knockout) significantly enhanced β-carotene production. A key mutant, YL19 (ΔHMO1), achieved a titer of 33.71 mg/L in shake-flasks─a 149.15% increase over the parent strain─with a concurrent lycopene reduction indicating redirected carbon flux toward β-carotene biosynthesis. This integrated platform enhances screening efficiency and provides a new paradigm for identifying critical terpenoid biosynthetic targets.

|
Scooped by
mhryu@live.com
March 7, 12:27 PM
|
Characterizing CRISPR interference (CRISPRi) phenotypes presents a fundamental temporal challenge: pre–existing overabundance of target proteins can mask early silencing, requiring extended growth for dilution, yet prolonged repression rapidly selects for escaper mutants. To resolve this, we integrated a tightly regulated CRISPRi system in Pseudomonas putida with an automated mini-bioreactor platform operating in turbidostat mode. By maintaining continuous exponential growth, we mapped the exact temporal dynamics of essential gene silencing. We identified a critical observation window between 17 and 27 hours (7–9.5 cell doublings) where repression exerts its maximum physiological impact, directly preceding population takeover by target-site mutated escapers. Applying this workflow to the arginine biosynthesis pathway, multi-omics profiling disentangled transient physiological buffering from long-term mutational events, and revealing that argH and argG knockdowns trigger highly diverse metabolomic perturbations. This scalable framework overcomes batch culture limitations, ensuring precise temporal control for accurate phenotypic characterization and reliable functional genomics.

|
Scooped by
mhryu@live.com
March 7, 12:18 PM
|
This study presents dAMN, a hybrid neural-mechanistic model that integrates neural networks with genome-scale dynamic flux balance analysis (dFBA) to predict bacterial growth curves across diverse nutrient environments. dAMN uses neural networks to infer dynamic behavior from initial metabolite concentrations, while mechanistic constraints ensure stoichiometric and thermodynamic consistency based on genome scale metabolic models. dAMN is trained on E. coli and P. putida experimental growth data from media containing various combinations of sugars, amino acids, and nucleobases, and evaluated on two test sets: one for forecasting over time and another for predicting growth dynamics on unseen media. dAMN achieved high predictive power (R > 0.9), successfully reproducing growth and substrate depletion dynamics including acetate overflow and glucose-acetate consumption shift for E. coli. An interesting innovation of dAMN is the treatment of the lag phase, enabling realistic adaptation dynamics absent from standard dFBA models. dAMN stands out for its ability to generalize across combinatorial nutrient inputs and produce full growth-curve predictions from minimal input data.

|
Scooped by
mhryu@live.com
March 7, 12:09 PM
|
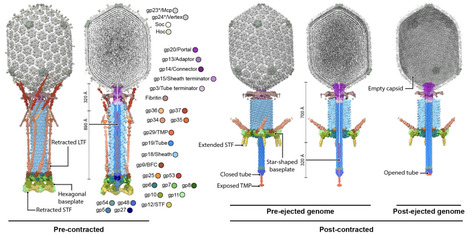
Successful viral infection requires efficient adsorption to the target cell, followed by membrane penetration for genome translocation into the host cytoplasm. Bacteriophage T4 initiates infection by recognising E. coli surface receptors via its long tail fibres. Receptor binding triggers sequential conformational changes that culminate in tail sheath contraction and genome delivery through viral channels spanning the host cell envelope. Despite extensive structural studies, the mechanism of genome translocation by tailed phages remains unclear. Here, using cryo-electron microscopy, we resolved structures of bacteriophage T4 at discrete stages. This revealed how long tail fibre extension affects the baseplate to initiate tail contraction, and the domain architecture of the tape measure protein and its involvement in channel formation and genome translocation. Furthermore, we demonstrate that the virus-encoded superinfection exclusion protein Imm binds to the tape measure protein to prevent secondary infections. Our findings reveal the mechanism of tape measure protein-mediated membrane penetration, offering insights into the coordinated process of genome delivery and phage-encoded superinfection exclusion proteins that prevent genome translocation.

|
Scooped by
mhryu@live.com
March 7, 11:57 AM
|
RNA interference (RNAi) shows great potential to protect agricultural crops against a variety of fungal diseases. However, protection efficiencies reported by empirical RNAi studies can vary greatly, and our understanding of the factors responsible for this variance remains limited. In this meta-analysis, we evaluated 89 studies that compare the efficiency of host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS) in controlling fungal diseases, focusing on biotrophic, hemibiotrophic, and necrotrophic fungi, the use of formulations and the dsRNA design as explanatory factors for differences between reported efficiency values. Our results indicate that SIGS is slightly more effective, particularly in biotrophs. Somewhat surprisingly, we did not find significant efficiency differences between SIGS studies that used formulations and those that applied naked dsRNA. We also considered the effect of various parameters describing RNA design. Differences in dsRNA length and the number of constructs, and number of targets showed no consistent significant effect on resistance in either HIGS or SIGS. Interestingly, however, HIGS studies reported significantly higher efficiency when targeting genes closer to the 3` end and SIGS when targeting genes closer to the 5` end. We discuss reasons for the reported patterns, such as variability in dsRNA uptake mechanisms, intercellular trafficking and Dicer processing, and conclude that more research is needed to understand the biological mechanisms determining RNAi efficiency for fungal control.

|
Scooped by
mhryu@live.com
March 7, 11:45 AM
|
Wood, once regarded primarily as a structural material, possesses rich physicochemical complexity that has long been underexplored. In the context of industrialization and carbon imbalance, it is now emerging as a renewable and multifunctional platform for green nano-technologies. Recent advances in wood nanotechnology have enabled the transformation of natural wood into programmable substrates with tailored nanoarchitectures, establishing it as a representative class of bio-based nanomaterials. This review systematically categorizes wood-specific nanoengineering strategies—including thermal carbonization, laser-induced graphenization, targeted delignification, nanomaterial integration, and mechanical processing—highlighting their mechanisms and impacts on wood’s multiscale structural and functional properties. Importantly, these functionalization strategies can be flexibly combined in a modular, “Lego-like” manner, enabling wood to be reconfigured and optimized for diverse application scenarios. We summarize recent progress in applying functionalized wood to sustainable technologies such as energy storage (e.g., metal-ion batteries, Zn–air systems, supercapacitors), water treatment (e.g., adsorption, photothermal filtration, catalytic degradation), and energy conversion (e.g., solar evaporation, ionic thermoelectrics, hydrovoltaics, and triboelectric nanogenerators). These studies reveal how nanoengineered wood structures can enable efficient charge transport, selective adsorption, and enhanced light-to-heat conversion. Finally, the review discusses current challenges—such as scalable fabrication, material integration, and long-term environmental stability—and outlines future directions for the development of wood-based platforms in next-generation green energy and environmental systems.

|
Scooped by
mhryu@live.com
March 7, 11:32 AM
|
Polyamines are ancient metabolites that serve critical functions in maintaining epithelial integrity, regulating immune response, and supporting healthy aging. The gut microbiota actively synthesizes and converts polyamines, while host factors such as inflammation, barrier function, and nutritional status dynamically modulate this metabolic network. Disruption of this host–microbiota axis reduces polyamine availability, impairs barrier function, and exacerbates inflammation. In contrast, polyamines exert protective effects by promoting epithelial repair, modulating macrophage and T-cell responses, and enhancing autophagy-mediated tissue renewal and longevity. Recent advances in engineered probiotics, microbial small RNAs, and postbiotics further highlight the therapeutic potential of precisely modulating polyamine metabolism in clinical contexts such as inflammatory bowel disease, metabolic syndrome, and neurodegenerative conditions associated with aging.
|
 Your new post is loading...
Your new post is loading...