Your new post is loading...

|
Scooped by
?
March 23, 2018 1:48 PM
|
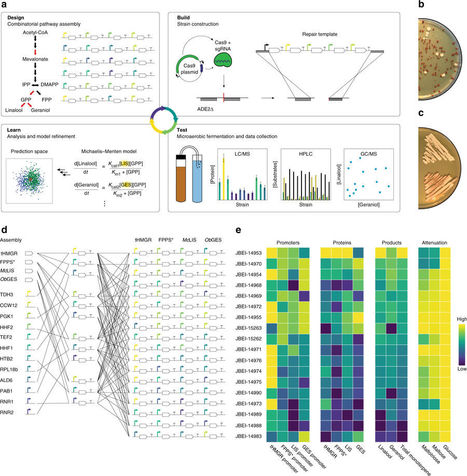
Flowers of the hop plant provide both bitterness and “hoppy” flavor to beer. Hops are, however, both a water and energy intensive crop and vary considerably in essential oil content, making it challenging to achieve a consistent hoppy taste in beer. Here, we report that brewer’s yeast can be engineered to biosynthesize aromatic monoterpene molecules that impart hoppy flavor to beer by incorporating recombinant DNA derived from yeast, mint, and basil. Whereas metabolic engineering of biosynthetic pathways is commonly enlisted to maximize product titers, tuning expression of pathway enzymes to affect target production levels of multiple commercially important metabolites without major collateral metabolic changes represents a unique challenge. By applying state-of-the-art engineering techniques and a framework to guide iterative improvement, strains are generated with target performance characteristics. Beers produced using these strains are perceived as hoppier than traditionally hopped beers by a sensory panel in a double-blind tasting.

|
Scooped by
?
March 20, 2018 11:38 AM
|
Increasing availability of new genomes and putative biosynthetic gene clusters (BGCs) has extended the opportunity to access novel chemical diversity for agriculture, medicine, environmental and industrial purposes. However, functional characterization of BGCs through heterologous expression is limited because expression may require complex regulatory mechanisms, specific folding or activation. We developed an integrated workflow for BGC characterization that integrates pathway identification, modular design, DNA synthesis, assembly and characterization. This workflow was applied to characterize multiple phenazine-modifying enzymes. Phenazine pathways are useful for this workflow because all phenazines are derived from a core scaffold for modification by diverse modifying enzymes (PhzM, PhzS, PhzH, and PhzO) that produce characterized compounds. We expressed refactored synthetic modules of previously uncharacterized phenazine BGCs heterologously in Escherichia coli and were able to identify metabolic intermediates they produced, including a previously unidentified metabolite. These results demonstrate how this approach can accelerate functional characterization of BGCs.

|
Scooped by
?
March 19, 2018 3:36 PM
|
An increasing number of studies have strongly correlated the composition of the human microbiota with many human health conditions and, in several cases, have shown that manipulating the microbiota directly affects health. These insights have generated significant interest in engineering indigenous microbiota community members and nonresident probiotic bacteria as biotic diagnostics and therapeutics that can probe and improve human health. In this review, we discuss recent advances in synthetic biology to engineer commensal and probiotic lactic acid bacteria, bifidobacteria, and Bacteroides for these purposes, and we provide our perspective on the future potential of these technologies.

|
Scooped by
?
March 19, 2018 8:32 AM
|
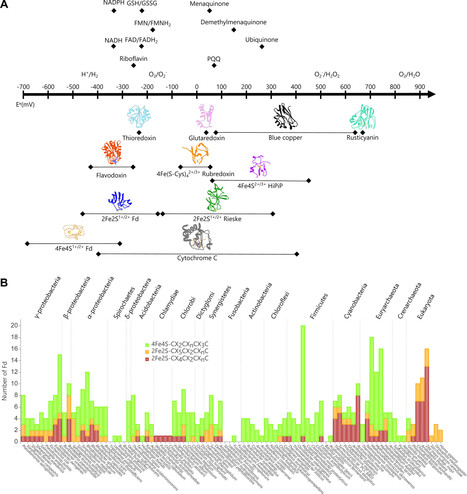
The ferredoxin (Fd) protein family is a structurally diverse group of iron–sulfur proteins that function as electron carriers, linking biochemical pathways important for energy transduction, nutrient assimilation, and primary metabolism. While considerable biochemical information about individual Fd protein electron carriers and their reactions has been acquired, we cannot yet anticipate the proportion of electrons shuttled between different Fd-partner proteins within cells using biochemical parameters that govern electron flow, such as holo-Fd concentration, midpoint potential (driving force), molecular interactions (affinity and kinetics), conformational changes (allostery), and off-pathway electron leakage (chemical oxidation). Herein, we describe functional and structural gaps in our Fd knowledge within the context of a sequence similarity network and phylogenetic tree, and we propose a strategy for improving our understanding of Fd sequence–function relationships. We suggest comparing the functions of divergent Fds within cells whose growth, or other measurable output, requires electron transfer between defined electron donor and acceptor proteins. By comparing Fd-mediated electron transfer with biochemical parameters that govern electron flow, we posit that models that anticipate energy flow across Fd interactomes can be built. This approach is expected to transform our ability to anticipate Fd control over electron flow in cellular settings, an obstacle to the construction of synthetic electron transfer pathways and rational optimization of existing energy-conserving pathways.

|
Scooped by
?
March 19, 2018 1:10 AM
|
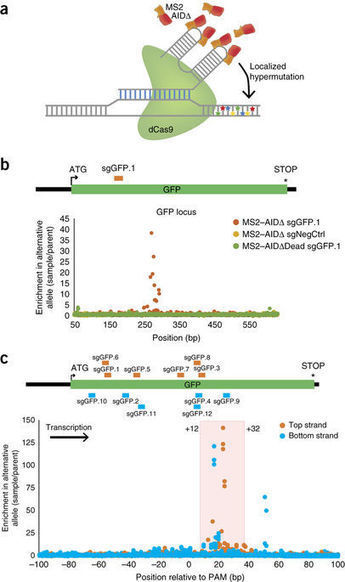
Engineering and study of protein function by directed evolution has been limited by the technical requirement to use global mutagenesis or introduce DNA libraries. Here, we develop CRISPR-X, a strategy to repurpose the somatic hypermutation machinery for protein engineering in situ. Using catalytically inactive dCas9 to recruit variants of cytidine deaminase (AID) with MS2-modified sgRNAs, we can specifically mutagenize endogenous targets with limited off-target damage. This generates diverse libraries of localized point mutations and can target multiple genomic locations simultaneously. We mutagenize GFP and select for spectrum-shifted variants, including EGFP. Additionally, we mutate the target of the cancer therapeutic bortezomib, PSMB5, and identify known and novel mutations that confer bortezomib resistance. Finally, using a hyperactive AID variant, we mutagenize loci both upstream and downstream of transcriptional start sites. These experiments illustrate a powerful approach to create complex libraries of genetic variants in native context, which is broadly applicable to investigate and improve protein function.

|
Scooped by
?
March 18, 2018 11:33 PM
|
Geological storage of CO2 is a fast-developing technology that can mitigate rising carbon emissions. However, there are environmental concerns with the long-term storage and implications of a leak from a carbon capture storage (CCS) site. Traditional monitoring lacks clear protocols and relies heavily on physical methods. Here, we discuss the potential of biotechnology, focusing on microbes with a natural ability to utilize and assimilate CO2 through different metabolic pathways. We propose the use of natural microbial communities for CCS monitoring and CO2 utilization, and, with examples, demonstrate how synthetic biology may maximize CO2 uptake within and above storage sites. An integrated physical and biological approach, combined with metagenomics data and biotechnological advances, will enhance CO2 sequestration and prevent large-scale leakages.

|
Scooped by
?
March 18, 2018 11:01 PM
|
The importance of the plant microbiome for host fitness has led to the concept of the “plant holobiont”. Seeds are reservoirs and vectors for beneficial microbes, which are very intimate partners of higher plants with the potential to connect plant generations. In this study, the endophytic seed microbiota of numerous barley samples, representing different cultivars, geographical sites and harvest years, was investigated. Cultivation–dependent and −independent analyses, microscopy, functional plate assays, greenhouse assays and functional prediction were used, with the aim of assessing the composition, stability and function of the barley seed endophytic bacterial microbiota. Associations were consistently detected in the seed endosphere with Paenibacillus, Pantoea and Pseudomonas spp., which were able to colonize the root with a notable rhizocompetence after seed germination. In greenhouse assays, enrichment with these bacteria promoted barley growth, improved mineral nutrition and induced resistance against the fungal pathogen Blumeria graminis. It was demonstrated that barley, an important crop plant, was consistently associated with beneficial bacteria inside the seeds. The results have relevant implications for plant microbiome ecology and for the holobiont concept, as well as opening up new possibilities for research and application of seed endophytes as bioinoculants in sustainable agriculture.

|
Scooped by
?
March 18, 2018 10:50 AM
|
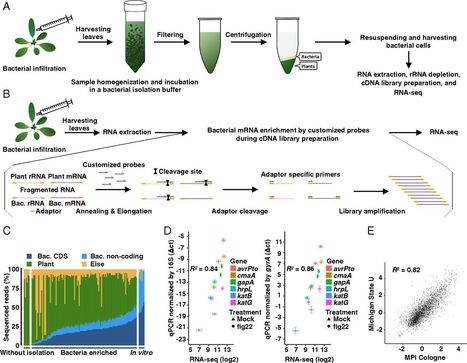
Plant pathogens can cause serious diseases that impact global agriculture. The plant innate immunity, when fully activated, can halt pathogen growth in plants. Despite extensive studies into the molecular and genetic bases of plant immunity against pathogens, the influence of plant immunity in global pathogen metabolism to restrict pathogen growth is poorly understood. Here, we developed RNA sequencing pipelines for analyzing bacterial transcriptomes in planta and determined high-resolution transcriptome patterns of the foliar bacterial pathogen Pseudomonas syringae in Arabidopsis thaliana with a total of 27 combinations of plant immunity mutants and bacterial strains. Bacterial transcriptomes were analyzed at 6 h post infection to capture early effects of plant immunity on bacterial processes and to avoid secondary effects caused by different bacterial population densities in planta . We identified specific “immune-responsive” bacterial genes and processes, including those that are activated in susceptible plants and suppressed by plant immune activation. Expression patterns of immune-responsive bacterial genes at the early time point were tightly linked to later bacterial growth levels in different host genotypes. Moreover, we found that a bacterial iron acquisition pathway is commonly suppressed by multiple plant immune-signaling pathways. Overexpression of a P. syringae sigma factor gene involved in iron regulation and other processes partially countered bacterial growth restriction during the plant immune response triggered by AvrRpt2. Collectively, this study defines the effects of plant immunity on the transcriptome of a bacterial pathogen and sheds light on the enigmatic mechanisms of bacterial growth inhibition during the plant immune response.

|
Scooped by
?
March 16, 2018 12:11 AM
|
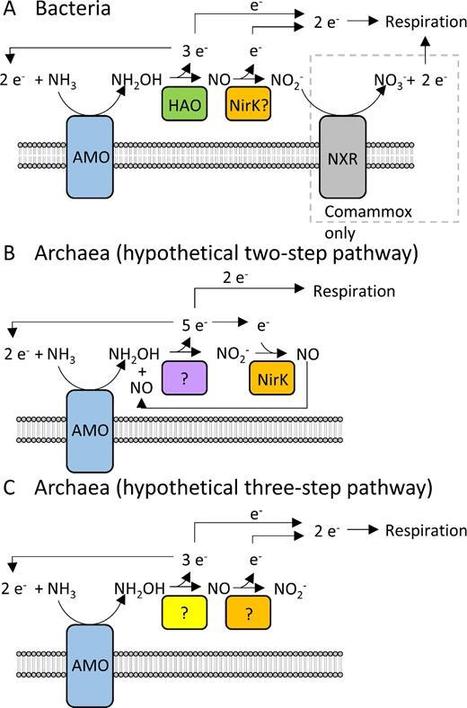
Ammonia oxidation is a fundamental core process in the global biogeochemical nitrogen cycle. Oxidation of ammonia (NH3) to nitrite (NO2−) is the first and rate-limiting step in nitrification and is carried out by distinct groups of microorganisms. Ammonia oxidation is essential for nutrient turnover in most terrestrial, aquatic and engineered ecosystems and plays a major role, both directly and indirectly, in greenhouse gas production and environmental damage. Although ammonia oxidation has been studied for over a century, this research field has been galvanised in the past decade by the surprising discoveries of novel ammonia oxidising microorganisms. This review reflects on the ammonia oxidation research to date and discusses the major gaps remaining in our knowledge of the biology of ammonia oxidation.

|
Scooped by
?
March 14, 2018 11:38 PM
|
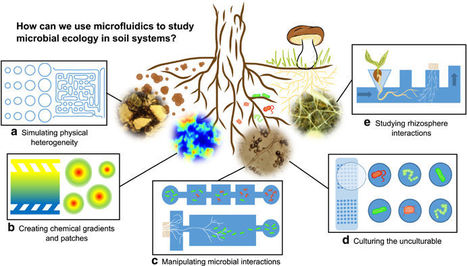
Microfluidic platforms have the potential to address four major challenges in studying soil systems, namely (1) their enormous spatio-temporal heterogeneity, (2) the lack of current methods to mimic soil realistically at the appropriate scale, (3) the difficulties in studying interactions among soil microbes and (4) the lack of optical access in real soil systems. We discuss these issues in turn, highlighting important advances driving the transfer of this technology to soil systems, but also identify potential challenges.

|
Scooped by
?
March 14, 2018 9:03 AM
|
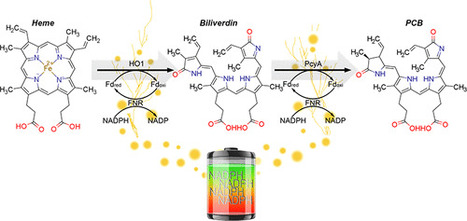
Transplanting metabolic reactions from one species into another has many uses as a research tool with applications ranging from optogenetics to crop production. Ferredoxin (Fd), the enzyme that most often supplies electrons to these reactions, is often overlooked when transplanting enzymes from one species to another because most cells already contain endogenous Fd. However, we have shown that the production of chromophores used in Phytochrome B (PhyB) optogenetics is greatly enhanced in mammalian cells by expressing bacterial and plant Fds with ferredoxin-NADP+ reductases (FNR). We delineated the rate limiting factors and found that the main metabolic precursor, heme, was not the primary limiting factor for producing either the cyanobacterial or plant chromophores, phycocyanobilin or phytochromobilin, respectively. In fact, Fd is limiting, followed by Fd+FNR and finally heme. Using these findings, we optimized the PCB production system and combined it with a tissue penetrating red/far-red sensing PhyB optogenetic gene switch in animal cells. We further characterized this system in several mammalian cell lines using red and far-red light. Importantly, we found that the light-switchable gene system remains active for several hours upon illumination, even with a short light pulse, and requires very small amounts of light for maximal activation. Boosting chromophore production by matching metabolic pathways with specific ferredoxin systems will enable the unparalleled use of the many PhyB optogenetic tools and has broader implications for optimizing synthetic metabolic pathways.

|
Scooped by
?
March 14, 2018 2:06 AM
|
We are in the midst of a major shift in how we think about treating disease and the regeneration of damaged tissues. Although small-molecule and protein therapeutics are the dominant forms of treatment today, we are now at the point where we can engineer our own body's cells to detect and treat disease. Using the cutting-edge tools of synthetic biology, we may one day be able to build smart therapeutic cells that reside in the body for life, poised to respond to diseases that would otherwise thwart our natural immune system. My colleagues and I envision cell therapies that act as microscopic “physicians,” capable of detecting, diagnosing, and directly eradicating disease via a multifaceted mechanism that is difficult to resist and circumvent. This therapeutic vision contrasts with more traditional drug therapies, which often require chronic administration and generally target individual disease mechanisms that are easily bypassed, resulting in disease recurrence.

|
Scooped by
?
March 13, 2018 11:01 PM
|
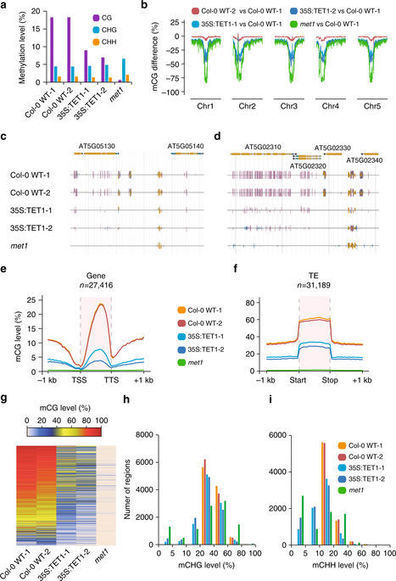
epimutagenesis introduces random methylation variation via the introduction of a transgeneepimutagenesis introduces random methylation variation via the introduction of a transgeneDNA methylation in the promoters of plant genes sometimes leads to transcriptional repression, and the loss of DNA methylation in methyltransferase mutants results in altered gene expression and severe developmental defects. However, many cases of naturally occurring DNA methylation variations have been reported, whereby altered expression of differentially methylated genes is responsible for agronomically important traits. The ability to manipulate plant methylomes to generate epigenetically distinct individuals could be invaluable for breeding and research purposes. Here, we describe “epimutagenesis,” a method to rapidly generate DNA methylation variation through random demethylation of the Arabidopsis thaliana genome. This method involves the expression of a human ten–eleven translocation (TET) enzyme, and results in widespread hypomethylation that can be inherited to subsequent generations, mimicking mutants in the maintenance of DNA methyltransferase met1. Application of epimutagenesis to agriculturally significant plants may result in differential expression of alleles normally silenced by DNA methylation, uncovering previously hidden phenotypic variations.
|

|
Scooped by
?
March 20, 2018 11:00 PM
|
A powerful contributor to prokaryotic evolution is horizontal gene transfer (HGT) through transformation, conjugation, and transduction, which can be advantageous, neutral, or detrimental to fitness. Bacteria and archaea control HGT and phage infection through CRISPR-Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) adaptive immunity. Although the benefits of resisting phage infection are evident, this can come at a cost of inhibiting the acquisition of other beneficial genes through HGT. Despite the ability of CRISPR-Cas to limit HGT through conjugation and transformation, its role in transduction is largely overlooked. Transduction is the phage-mediated transfer of bacterial DNA between cells and arguably has the greatest impact on HGT. We demonstrate that in Pectobacterium atrosepticum, CRISPR-Cas can inhibit the transduction of plasmids and chromosomal loci. In addition, we detected phage-mediated transfer of a large plant pathogenicity genomic island and show that CRISPR-Cas can inhibit its transduction. Despite these inhibitory effects of CRISPR-Cas on transduction, its more common role in phage resistance promotes rather than diminishes HGT via transduction by protecting bacteria from phage infection. This protective effect can also increase transduction of phage-sensitive members of mixed populations. CRISPR-Cas systems themselves display evidence of HGT, but little is known about their lateral dissemination between bacteria and whether transduction can contribute. We show that, through transduction, bacteria can acquire an entire chromosomal CRISPR-Cas system, including cas genes and phage-targeting spacers. We propose that the positive effect of CRISPR-Cas phage immunity on enhancing transduction surpasses the rarer cases where gene flow by transduction is restricted.

|
Scooped by
?
March 19, 2018 9:58 PM
|
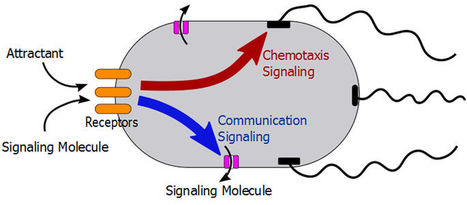
Bacteria are able to coordinate their movement, growth and biochemical activities through cell-cell communication. While the biophysical mechanism of bacterial chemotaxis has been well understood in individual cells, the role of communication in the chemotaxis of bacterial populations is not clear. Here we report experimental evidence for cell-cell communication that significantly enhances the chemotactic migration of bacterial populations, a finding that we further substantiate using numerical simulations. Using a microfluidic approach, we find that E. coli cells respond to the gradient of chemoattractant not only by biasing their own random-walk swimming pattern through the well-understood intracellular chemotaxis signaling, but also by actively secreting a chemical signal into the extracellular medium, possibly through a hitherto unknown communication signal transduction pathway. This extracellular signaling molecule is a strong chemoattractant that attracts distant cells to the food source. The observed behavior may represent a common evolved solution to accelerate the function of biochemical networks of interacting cells.

|
Scooped by
?
March 19, 2018 8:38 AM
|
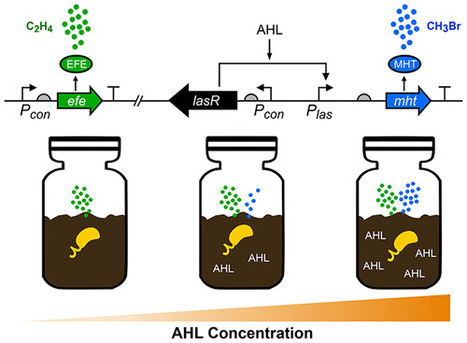
Fluorescent proteins are ubiquitous tools that are used to monitor the dynamic functions of natural and synthetic genetic circuits. However, these visual reporters can only be used in transparent settings, a limitation that complicates nondisruptive measurements of gene expression within many matrices, such as soils and sediments. We describe a new ratiometric gas reporting method for nondisruptively monitoring gene expression within hard-to-image environmental matrices. With this approach, C2H4 is continuously synthesized by ethylene forming enzyme to provide information on viable cell number, and CH3Br is conditionally synthesized by placing a methyl halide transferase gene under the control of a conditional promoter. We show that ratiometric gas reporting enables the creation of Escherichia coli biosensors that report on acylhomoserine lactone (AHL) autoinducers used for quorum sensing by Gram-negative bacteria. Using these biosensors, we find that an agricultural soil decreases the bioavailable concentration of a long-chain AHL up to 100-fold. We also demonstrate that these biosensors can be used in soil to nondisruptively monitor AHLs synthesized by Rhizobium leguminosarum and degraded by Bacillus thuringiensis. Finally, we show that this new reporting approach can be used in Shewanella oneidensis, a bacterium that lives in sediments.

|
Scooped by
?
March 19, 2018 8:30 AM
|
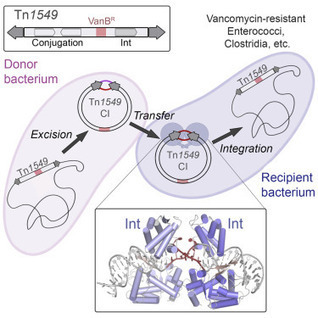
Conjugative transposition drives the emergence of multidrug resistance in diverse bacterial pathogens, yet the mechanisms are poorly characterized. The Tn1549 conjugative transposon propagates resistance to the antibiotic vancomycin used for severe drug-resistant infections. Here, we present four high-resolution structures of the conserved Y-transposase of Tn1549 complexed with circular transposon DNA intermediates. The structures reveal individual transposition steps and explain how specific DNA distortion and cleavage mechanisms enable DNA strand exchange with an absolute minimum homology requirement. This appears to uniquely allow Tn916-like conjugative transposons to bypass DNA homology and insert into diverse genomic sites, expanding gene transfer. We further uncover a structural regulatory mechanism that prevents premature cleavage of the transposon DNA before a suitable target DNA is found and generate a peptide antagonist that interferes with the transposase-DNA structure to block transposition. Our results reveal mechanistic principles of conjugative transposition that could help control the spread of antibiotic resistance genes.

|
Scooped by
?
March 18, 2018 11:37 PM
|
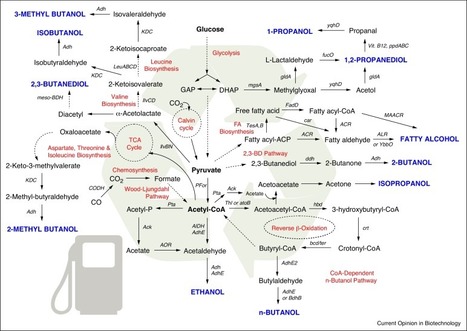
Bioengineering and synthetic biology approaches have revolutionised the field of biotechnology, enabling the introduction of non-native and de novo pathways for biofuels production. This ‘retooling’ of microorganisms is also applied to the utilisation of mixed carbon components derived from lignocellulosic biomass, a major technical barrier for the development of economically viable fermentations. This review will discuss recent advances in microorganism engineering for efficient production of alcohols from waste biomass. These advances span the introduction of new pathways to alcohols, host modifications for more cost-effective utilisation of lignocellulosic waste and modifications of existing pathways for generating new fuel additives.

|
Scooped by
?
March 18, 2018 11:26 PM
|
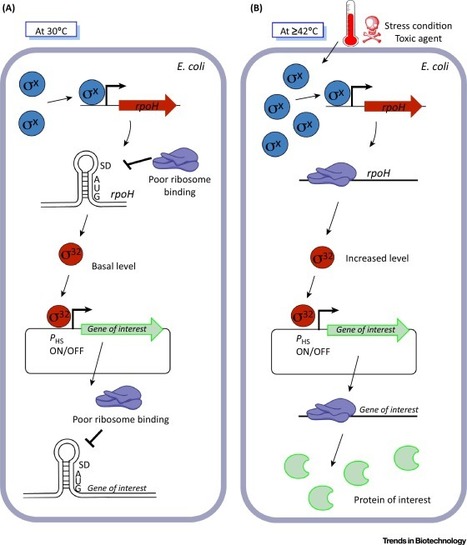
The Escherichia coli heat shock response (HSR) is a complex mechanism triggered by heat shock and by a variety of other growth-impairing stresses. We explore here the potential use of the E. coli HSR mechanism in synthetic biology approaches. Several components of the regulatory mechanism (such as heat shock promoters, proteins, and RNA thermosensors) can be extremely valuable in the creation of a toolbox of well-characterized biological parts to construct biosensors or microbial cell factories with applications in the environment, industry, or healthcare. In the future, these systems can be used for instance to detect a pollutant in water, to regulate and optimize the production of a compound with industrial relevance, or to administer a therapeutic agent in vivo.

|
Scooped by
?
March 18, 2018 10:33 PM
|
Microbes typically live alongside many other species in complex communities. These microbial communities are very important for us because they also live in and on our bodies and can determine our health and well-being. The composition and function of these communities, such as who is part of such a community and who is excluded, are decided by the interactions between the microbes. These microbial interactions can be driven by many different factors such as resource competition or toxin production. Although these factors are all different, the interactions are typically mediated through the environment; the microbes modify the environment, and they and other microbes have to live in this new environment. We show here that by understanding how microbes change and react to the environment, it is possible to understand and even predict their interactions. We believe that this way of thinking about microbial interactions will lead to a better understanding of more complex communities that are so important for our well-being.

|
Scooped by
?
March 16, 2018 9:57 AM
|
Digested genome sequencing, or Digenome-seq, is an in vitro assay that has become increasingly popular since its introduction in 2015. It has a simple two-step protocol: in vitro Cas9 cleavage followed by next-gen sequencing. Two newer methods, CIRCLE-Seq and SITE-Seq, are slightly more complex but are also more sensitive, as they enrich for nuclease-cleaved genomic DNA before sequencing. Researchers are continuing to improve the accuracy and throughput of these methods. “I think in the long term, in vitro is going to be the way to go, but that’s an evolving space right now,” says Joung.

|
Scooped by
?
March 15, 2018 11:59 PM
|
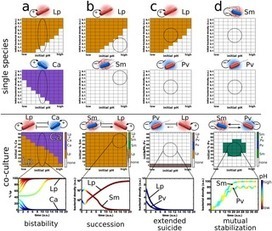
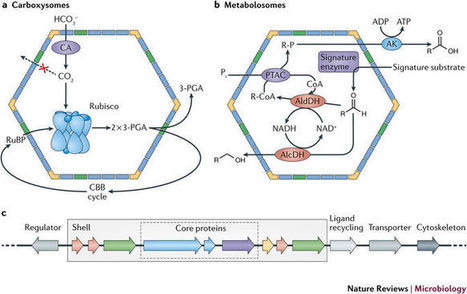
*Bacterial microcompartments are functional analogues of the lipid-bound organelles of eukaryotes. They enclose chemical reactions that benefit from being separated from the cytosol. The delimiting membrane of bacterial microcompartments consists entirely of protein, and its components are highly conserved in sequence and structure. Bacterial microcompartments are found in a wide variety of bacterial species (at least 19 established phyla). They are easily identified in genomes by their tendency to colocalize the associated genes into a large gene cluster called a superlocus. Carboxysomes (CO2-fixing organelles) were the first type of bacterial microcompartment to be identified, but recently, many more have been discovered and characterized; they are involved in catabolizing a variety of nutrients and enable cells to grow in otherwise unavailable niches. The shell and cargo of bacterial microcompartments self-assemble using different pathways; some build the shell around a cargo aggregate, whereas others assemble the shell and cargo concomitantly. There are proteins that facilitate cargo aggregation and small encapsulation peptides that specifically associate proteins to the lumen of the shell. Bacterial microcompartments are linked to the pathogenesis of certain bacteria because they confer a growth advantage. For example, the human gut is enriched in propanediol and ethanolamine, initial substrates of specific bacterial microcompartments. The knowledge gained from understanding the native functions has led to substantial progress in modifying the shell for bioengineering purposes. Bacterial microcompartment shells can be produced recombinantly, and shell proteins and cores have been engineered to adopt new functions.

|
Scooped by
?
March 14, 2018 11:26 PM
|
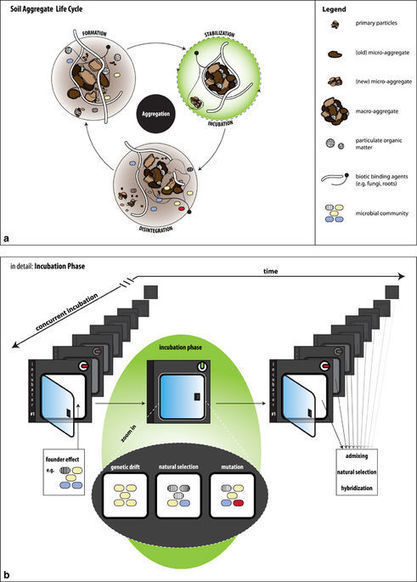
Soil aggregation, a key component of soil structure, has mostly been examined from the perspective of soil management and the mediation of ecosystem processes such as soil carbon storage. However, soil aggregation is also a major factor to consider in terms of the fine-scale organization of the soil microbiome. For example, the physico-chemical conditions inside of aggregates usually differ from the conditions prevalent in the bulk soil and aggregates therefore increase the spatial heterogeneity of the soil. In addition, aggregates can provide a refuge for microbes against predation since their interior is not accessible to many predators. Soil aggregates are thus clearly important for microbial community ecology in soils and for microbially driven biogeochemistry, and soil microbial ecologists are increasingly appreciating these aspects of soil aggregation. Soil aggregates have, however, so far been neglected when it comes to evolutionary considerations and we here propose that the process of soil aggregation should be considered as an important driver of evolution in the soil microbial community.

|
Scooped by
?
March 14, 2018 2:10 AM
|
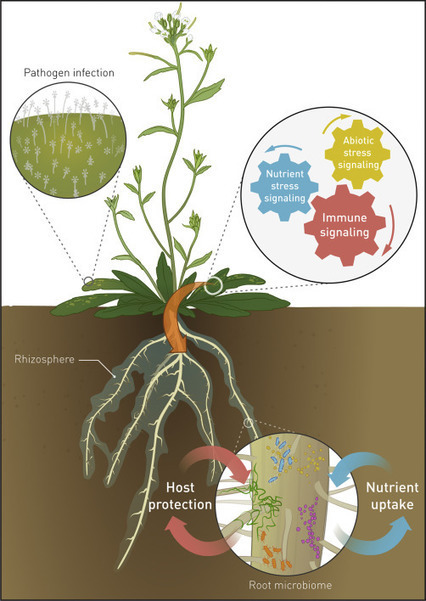
The rhizosphere microbiome extends the functional repertoire of plants beyond imagination. Exciting developments in high-throughput molecular analyses of the plant microbiome pinpointed enhanced nutrient uptake, improvement of root architecture, and protection of the host against biotic and abiotic stress as key functions of the microbiome. Currently, the life processes that shaped the co-evolution between plant hosts and their beneficial microbiota members gradually come to surface. It appears that plants evolved adaptive strategies by which they utilize root-associated microbiota to optimize both nutrient acquisition and immunity. Knowledge on the mechanisms and plant genetic pathways involved provides great potential for sustainable microbiome-based improvements of our future crops.

|
Scooped by
?
March 14, 2018 1:15 AM
|
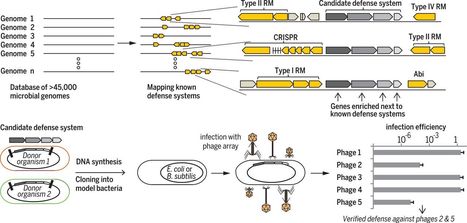
The arms race between bacteria and phages led to the development of sophisticated antiphage defense systems, including CRISPR-Cas and restriction-modification systems. Evidence suggests that known and unknown defense systems are located in “defense islands” in microbial genomes. Here, we comprehensively characterized the bacterial defensive arsenal by examining gene families that are clustered next to known defense genes in prokaryotic genomes. Candidate defense systems were systematically engineered and validated in model bacteria for their antiphage activities. We report nine previously unknown antiphage systems and one antiplasmid system that are widespread in microbes and strongly protect against foreign invaders. These include systems that adopted components of the bacterial flagella and condensin complexes. Our data also suggest a common, ancient ancestry of innate immunity components shared between animals, plants, and bacteria.
|
 Your new post is loading...
Your new post is loading...