Your new post is loading...

|
Scooped by
?
Today, 9:56 AM
|
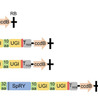
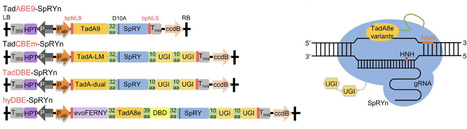
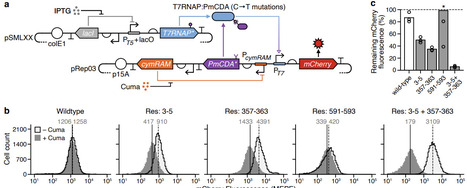
Base editing enables precise nucleotide substitutions within a relatively broad editing window (5–6 nucleotides). However, considerable bystander editing significantly compromise its accuracy. Point mutagenesis, a powerful approach for gradient-tuning protein function, facilitates the generation of diverse plant phenotypes to meet the demands of complex environments and consumer preferences. Here, a series of plant base editors is engineered by fusing three optimized TadA8e variants, TadA9, TadA-LM, and TadA-dual, with a PAM-flexible SpRY nickase (SpRYn, with 5′-NNN PAM recognition). These editors enable A-to-G, C-to-T, and dual-base (simultaneous A-to-G and C-to-T) conversions within a highly condensed active window (1–3 nucleotides). Performance evaluations reveal that the TadDBE (TadA Dual-Base Editor) achieves the most robust outcomes, delivering dual-base editing efficiencies ranging from 2.3% to 61.4%, while maintaining minimal off-target activity. Utilizing TadDBE, targeted point mutagenesis is performed on OsBadh2, a gene encoding betaine aldehyde dehydrogenase that plays a critical role in the biosynthesis of 2-acetyl-1-pyrroline (2-AP), a key aromatic compound. This approach yields rice lines exhibiting gradient-tuned aromatic profiles and optimized levels of 2-AP and γ-aminobutyric acid (GABA). These evolved TadA-derived editors provide a precise, PAM-flexible platform for base editing and represent a versatile strategy for generating genome-edited plants with gradient-tuned agronomic traits.

|
Scooped by
?
Today, 9:31 AM
|
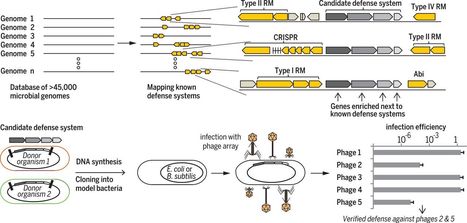
Risk assessment frameworks for plant agricultural biotechnology products have been in place for decades, focused on the evaluation of living biotechnology products created through genetic engineering. These products contain genetic material from outside the breeder’s gene pool, which is often from different taxa or represents “novel combinations of genetic material”. These products are typically considered to be “genetically modified” (GM) organisms in regulatory jurisdictions. However, in the microbial world, particularly among Bacteria and Archaea, the rapid expansion of genome sequence databases shows that natural microbial innovation primarily occurs through the natural exchange of genetic material from various sources, even from different taxa. This means that many microbes can be considered naturally occurring GM organisms. This raises the question of whether labeling a microbe as GM is always scientifically relevant for risk assessment. In most regulatory frameworks, being classified as GM significantly impacts the registration path, especially for microbes intended for environmental release. A more effective and science-based regulatory approach would assess the actual functions of a microbe rather than relying on the uncertain classification of its genetic material. This would benefit regulators, developers, and society by promoting the use of microbial technologies for agricultural use.

|
Scooped by
?
Today, 1:02 AM
|
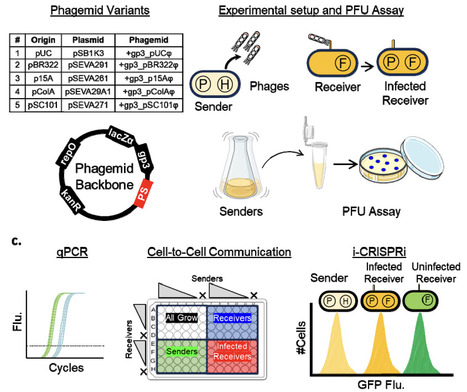
Intercellular communication is essential for distributed genetic circuits operating across cells in multicellular consortia. While diverse signalling molecules have been employed--ranging from quorum sensing signals, secondary metabolites, and pheromones to peptides, and nucleic acids--phage-packaged DNA offers a highly programmable method for communicating information between cells. Here, we present a library of five M13 phagemid variants with distinct replication origins, including those based on the Standard European Vector Architecture (SEVA) family, designed to tune the growth and secretion dynamics of sender strains. We systematically characterize how intracellular phagemid copy number varies with cellular growth physiology and how this, in turn, affects phage secretion rates. In co-cultures, these dynamics influence resource competition and modulate communication outcomes between sender and receiver cells. Leveraging the intercellular CRISPR interference (i-CRISPRi) system, we quantify phagemid transfer frequencies and identify rapid-transfer variants that enable efficient, low-burden communication. The phagemid toolbox developed here expands the repertoire of available phagemids for DNA-payload delivery applications and for implementing intercellular communication in multicellular circuits.

|
Scooped by
?
Today, 12:28 AM
|
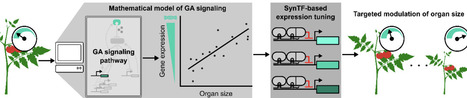
Enhancements to crop morphology, such as the semi-dwarfing that helped drive the green revolution, are often driven by changes in gene dosage. These changes are challenging to translate across varieties and species, which slows the pace of crop improvement. Synthetic transcription factors (SynTFs) offer a rapid alternative to generate targeted alterations to gene dosage. However, the complexity of developmental pathways makes it unclear how to best apply them to predictably engineer morphology. In this work, we explore if mathematical modeling can guide SynTF-based expression modulation of genes in the signaling pathway of the phytohormone, gibberellin (GA), which is a central regulator of cell expansion, to elucidate the design principles for engineering organ size. We demonstrate that modulation of GA signaling gene expression can generate consistent dwarfing across tissues in both controlled and variable environments in the model plant Arabidopsis thaliana, and that the degree of dwarfing is dependent on the strength of regulation as predicted by modeling. We further validate the predictive power of the model by demonstrating its capacity to accurately predict the qualitative impacts of different regulatory architectures for both increasing and decreasing organ size. Finally, we show that these insights can be generalized for engineering organ size in the crop Solanum lycopersicum (tomato). This work creates a framework for predictable engineering of an agriculturally important trait, organ size, across tissues and plant species. It also serves as a proof-of-concept for how mathematical models can guide SynTF-based alterations in gene dosage to enable bottom-up design of plant phenotypes.

|
Scooped by
?
June 22, 10:49 PM
|
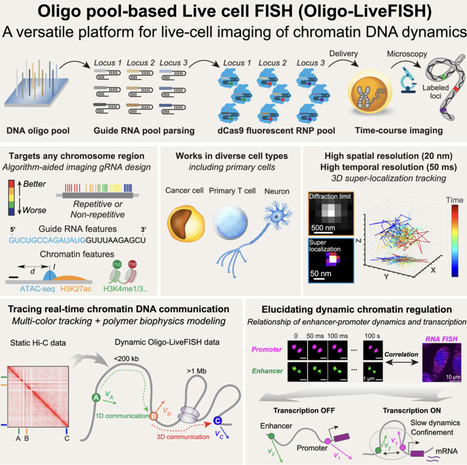
Three-dimensional (3D) genome dynamics are crucial for cellular functions and disease. However, real-time, live-cell DNA visualization remains challenging, as existing methods are often confined to repetitive regions, suffer from low resolution, or require complex genome engineering. Here, we present Oligo-LiveFISH, a high-resolution, reagent-based platform for dynamically tracking non-repetitive genomic loci in diverse cell types, including primary cells. Oligo-LiveFISH utilizes fluorescent guide RNA (gRNA) oligo pools generated by computational design, in vitro transcription, and chemical labeling, delivered as ribonucleoproteins. Utilizing machine learning, we characterized the impact of gRNA design and chromatin features on imaging efficiency. Multi-color Oligo-LiveFISH achieved 20-nm spatial resolution and 50-ms temporal resolution in 3D, capturing real-time enhancer and promoter dynamics. Our measurements and dynamic modeling revealed two distinct modes of chromatin communication, and active transcription slows enhancer-promoter dynamics at endogenous genes like FOS. Oligo-LiveFISH offers a versatile platform for studying 3D genome dynamics and their links to cellular processes and disease.

|
Scooped by
?
June 22, 2:49 PM
|
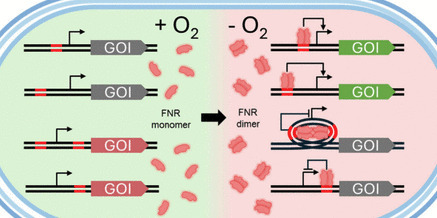
Promoters responsive to changes in cultivation conditions are essential tools for dynamic metabolic engineering. Oxygen-responsive promoters, in particular, exhibit significant application potential in oxygen-limited fermentation processes. However, currently reported oxygen-dependent promoters exhibit limited dynamic ranges, and notably, there remains a lack of research on oxygen-responsive negatively regulated promoters. In this study, we designed and characterized a series of dissolved oxygen-responsive promoters in Escherichia coli under the regulation of the transcription factor fumarate-nitrate reduction (FNR). Anaerobically activated promoters were constructed by inserting FNR binding site (FBS) upstream of inducible core promoters, while anaerobically repressed promoters were developed by inserting FBS downstream of or flanking constitutive promoters. The most effective anaerobically activated promoters showed 24–138-fold higher activity under anaerobic conditions compared to aerobic conditions. Under anaerobic conditions, promoters with DNA looping-mediated anaerobic repression maintained only 8–17% of the activity observed under aerobic conditions. These promoters were specifically regulated by FNR, as confirmed by tests in a DH5α Δfnr strain, and responded rapidly to oxygen depletion (within 30 min). The utility of these genetic tools was demonstrated by applying them to enhance pyruvate production in E. coli. An engineered strain with anaerobic-repressed aceE and anaerobic-activated atpAGD genes produced 5.76 g/L pyruvate at 55.7% yield in shake flask fermentations. This study offers an expanded toolbox of oxygen-responsive promoters that enable precise gene regulation based on dissolved oxygen levels, providing novel genetic strategies for developing efficient two-stage fermentation processes with separated growth and production phases.

|
Scooped by
?
June 22, 2:44 PM
|
RNA design has emerged to play a crucial role in synthetic biology and therapeutics. Although tertiary structure-based RNA design methods have been developed recently, they still overlook the broader molecular context, such as interactions with proteins, ligands, DNA, or ions, limiting the accuracy and functionality of designed sequences. To address this challenge, we present RISoTTo (RIbonucleic acid Sequence design from TerTiary structure), a parameter-free geometric deep learning approach that generates RNA sequences conditioned on both their backbone scaffolds and the surrounding molecular context. We evaluate the designed sequences based on their native sequence recovery rate and further validate them by predicting their secondary structures in silico and comparing them to the corresponding native structures. RISoTTo performs well on both metrics, demonstrating its ability to generate accurate and structurally consistent RNA sequences. Additionally, we present an in silico design study of domain 1 of the NAD+ riboswitch, where RISoTTo generated sequences are predicted to exhibit enhanced binding affinity for both the U1A protein and the NAD+ ligand.

|
Scooped by
?
June 22, 2:32 PM
|
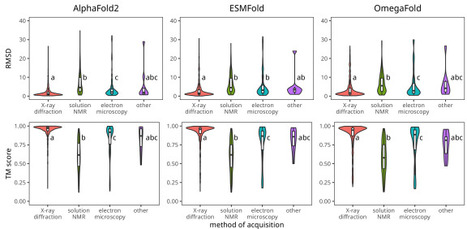
We compared the performance of three widely used protein structure prediction tools - AlphaFold2, ESMFold, and OmegaFold - using a dataset of over 1,300 newly created records from the PDB database. These structures, resolved between July 2022 and July 2024, ensure unbiased evaluation, as they were unavailable during the training of these tools. Using metrics such as root mean square deviation (RMSD), template modeling score (TM-score), and predicted local distance difference test (pLDDT), we found that AlphaFold2 consistently achieves the highest accuracy but depends on high-quality sequence alignments. In contrast, ESMFold and OmegaFold provide faster predictions and excel in challenging cases, such as rapidly evolving or designed proteins with limited sequence homology. Comparing ESMFold and OmegaFold, ESMFold achieves higher confidence scores (pLDDT) and structural similarity (TM-score). OmegaFold is competitive in specific contexts, such as de novo-designed proteins or sequences with limited evolutionary information. Additionally, we demonstrate that machine learning models trained on protein language model embeddings and pLDDT confidence scores can predict potential structure prediction failures, helping to identify challenging cases early in the pipeline.

|
Scooped by
?
June 22, 2:25 PM
|
The scientific study of animal venoms covers a broad phylogenetic domain. We argue that the true extent of this domain has been obscured by researchers having overlooked the biological essence of venom. Venoms manipulate the physiological functioning of recipients to produce extended phenotypes that are beneficial to the venom producer and detrimental to its victim. The ability to produce extended phenotypes in living victims, such as prey paralysis, distinguishes venom from saliva. Understanding venom from this perspective substantially broadens the phylogenetic domain of venom to include taxa that use toxic secretions to feed on plants and manipulate sexual partners, and it paves the way for unifying the field of venomics with the fields that study invertebrate–plant interactions and sexual conflict

|
Scooped by
?
June 22, 1:36 PM
|
Synthetic biology can endow microorganisms with novel capabilities. Su et al. developed an engineered strain of Vibrio natriegens that efficiently degrades complex organic pollutants in high-salinity environments. This technology offers a new biotechnological solution to urgent global environmental challenges, such as petrochemical wastewater discharges and marine oil spills.

|
Scooped by
?
June 22, 1:28 PM
|
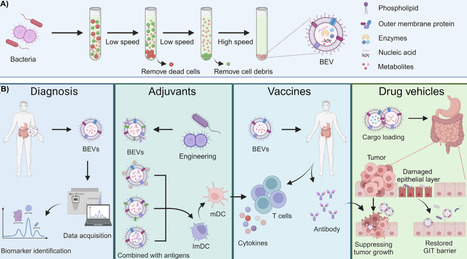
Bacterial extracellular vesicles (BEVs) are nanoscale spherical particles with lipid bilayer membranes containing diverse functional components from their parent bacteria. They exert pivotal effects on bacteria–bacteria and host–bacteria communication. Analyzing the dynamic changes in the production and composition of BEVs provides insights into the relationship between gut microbiota and host health, offering valuable perspectives for diagnosing various gastrointestinal diseases. Furthermore, BEVs can be employed as natural medications and drug delivery vehicles for inflammation management, cancer treatment, and vaccine development. This review summarizes the structural composition, generation mechanism, and biomedical applications of BEVs, emphasizing recent advances in immune regulation, gut microbiota modulation, and the clinical translation challenges associated with gastrointestinal diseases.

|
Scooped by
?
June 22, 1:25 PM
|
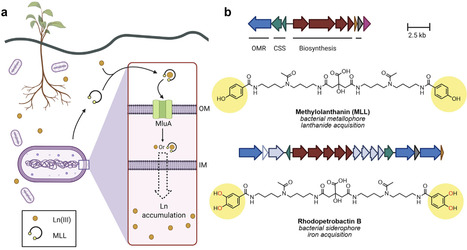
Metallophores are metal-chelating secondary metabolites that bacteria, fungi, and other organisms produce to acquire essential metal nutrients from their environment. This review highlights some recent discoveries in the field: the role of yersiniabactin in quorum sensing, the capacity of aspergillomarasmine A to inhibit bacterial Ni uptake, and the identification of methylolanthanin, a novel metallophore that enables bacterial lanthanide acquisition.

|
Scooped by
?
June 22, 1:07 PM
|
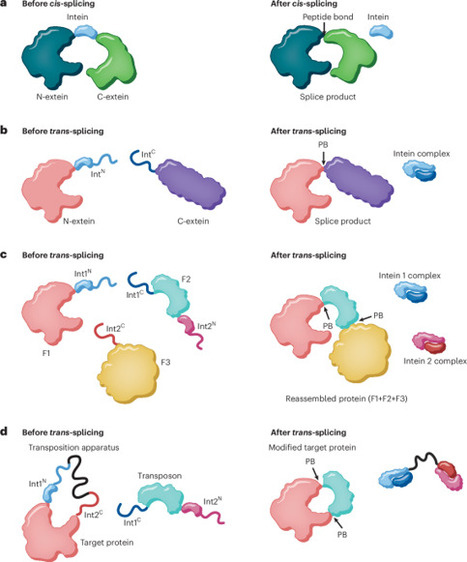
Transposition is normally considered a DNA-based process. Now, scientists have devised a means to carry out a similar transformation — this time using proteins. The approach enables protein semisynthesis and site-specific modification of folded substrates in vitro and in live cells, opening up new ways to probe and manipulate biological systems.
|

|
Scooped by
?
Today, 9:33 AM
|
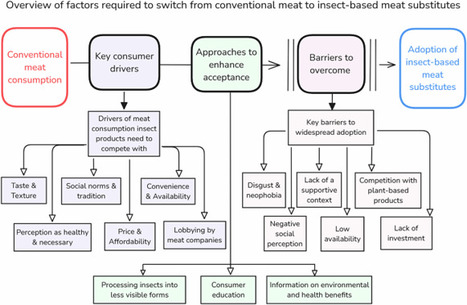
The substantial environmental footprint of meat production means that dietary shifts are needed to reduce greenhouse gas emissions. Insects may offer one alternative, but must first be widely accepted and consumed by the general public. This review evaluates the prospects of insect-based foods to compete with meat. We find that insect-based foods face major challenges, including low consumer acceptance and limited investment. They have a low likelihood of significantly reducing meat consumption, particularly when compared to more accepted plant-based alternatives.

|
Scooped by
?
Today, 8:45 AM
|
Escherichia coli, a key workhorse in industrial biotechnology, is commonly grown on glucose, which supports rapid growth but leads to acetate overflow, which instead inhibits growth, diverts carbon from the production pathway, and reduces productivity. Recent studies suggest that acetate may also have beneficial effects in glucose-grown E. coli, but its potential in bioprocesses remains unexplored. In this study, we systematically investigated acetate's impact on bioproduction using a kinetic model of glucose and acetate metabolism in E. coli. The model predicts that acetate can enhance bioproduction in glucose-grown E. coli through three mechanisms: (i) by minimizing acetate overflow, thereby reducing carbon loss, (ii) by increasing acetyl-CoA levels, thereby boosting the biosynthetic flux of acetyl-CoA-derived compounds, and (iii) by promoting biomass accumulation, thus improving overall productivity. We experimentally validated the predictions of the model for mevalonate and 3-hydroxypropionate production, where acetate supplementation increased productivity by 117% and 34%, respectively. Our findings provide a valuable framework for optimizing E. coli-based bioprocesses and highlight acetate's underutilized potential in biotechnology. By leveraging acetate from waste streams as a metabolic booster, this approach could contribute to more sustainable and environmentally friendly bioprocesses.

|
Scooped by
?
Today, 12:38 AM
|
Protein-protein conjugation systems are a powerful way of creating fusion proteins and enable the dynamic combination of protein domains with diverse functionalities. However, the insertion of these systems into enzymes is often performed with little consideration of the structural impact they might have. This is particularly relevant when modifying complex molecular machines that transition between numerous conformational states. Here, we address this issue by developing SIMPLIFE, a computational workflow that supports the design of optimal insertion sites for conjugation tags based on the structure of the proteins involved and performs localised residue redesign where needed. We demonstrate how SIMPLIFE can be used to effectively augment the function of T7 RNA polymerase using the DogCatcher-DogTag system, enabling diverse and dynamically varying mutations within a targeted region of DNA. This work demonstrates the power of combining biophysical and machine learning based approaches for protein structure prediction to efficiently augment the function of molecular machines, accelerating our ability to combine complex biochemical functionalities in new ways.

|
Scooped by
?
June 22, 11:36 PM
|
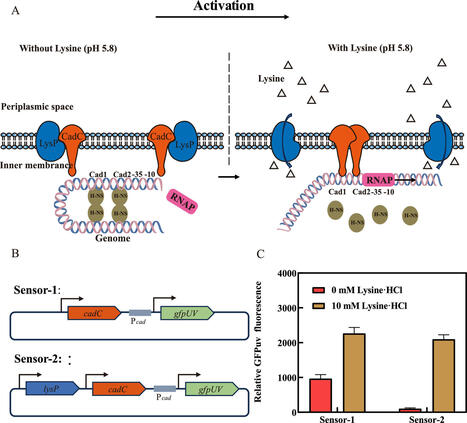
As one of the most promising monomers of biobased polyamides, cadaverine has wide industrial application prospects. However, in microbial cadaverine fermentation with glucose as the sole carbon source, the impaired coordination between precursor (lysine) utilization and cytotoxic cadaverine accumulation has been identified as the primary bottleneck limiting high-yield biosynthesis. Here, we developed a lysine biosensor in Escherichia coli to dynamically regulate cadaverine biosynthesis. Here, we developed a lysine biosensor based on the lysine transporter protein LysP, the transcription activator CadC, and the GFPuv gene under the control of the Pcad promoter. However, the engineered lysine biosensor system had a low dynamic range and a narrow pH operating range. Therefore, a multilevel optimization strategy, which included the introduction of key point mutations and engineered promoter modifications, were introduced to improve the performance of the biosensor, resulting in significant improvements in the dynamic range and lysine response. Moreover, we engineered a cadaverine-producing E. coli strain by increasing the supply of the lysine precursor, overexpressing key cadaverine synthesis genes, and knocking out genes related to metabolic bypass. The lysine biosensor was subsequently implemented to dynamically regulate cadaverine biosynthesis, resulting in a 48.10% increase in the production titre (33.19 g/L) and a 21.2% increase in cell growth compared with those resulting from the strain with constitutive expression. This is the first report in which a lysine biosensor constructed in E. coli could dynamically regulate cadaverine synthesis to improve its yield and biomass. This strategy provides new insights into the metabolic engineering of lysine and its derivatives in E. coli.

|
Scooped by
?
June 22, 10:45 PM
|
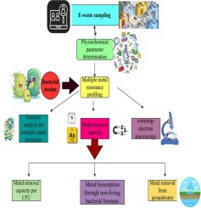
Samples were collected from two e-waste dumping sites (Mehmood Booti (31°36′28″N, 74°23′36″E) and Lakhodair (31°37′36.6″ N, 74°25′07.6″ E)) in Lahore, Pakistan. A portable multiparameter was used to determine physicochemical parameters such as temperature, pH, electrical conductivity, turbidity, total suspended particles, and total dissolved solids. Minimal salt broth was used for the determination of the minimal inhibitory concentration of the bacterium against all heavy metals. Bacterial morphology was observed under a scanning electron microscope with and without metal stress. The temperature range for all these samples was 28.7 to 35.7 °C, while the pH range was 6.7 to 7.89. The other parameters range, such as electrical conductivity µS/cm (698–8742), turbidity (14.2–103), total suspended particles (31–698), and total dissolved solids (564–23456). The lead concentration in the Mehmood Booti soil sample was 1800 mg/kg, while in the Lakhodair soil, it was 1567 mg/kg. Microbacterium sp. strain 1S1 was utilized for bioremediation assay at the lab and pilot scale. The resistance capacity of this bacterium against different metals was in the following order: As > Pb > Cd > Cu > Cr > Ni. The bioremediation potential of the bacterium against arsenic was 81.33 % and 96 % after 2 and 4 days. The least activity was observed against nickel, which was 17 and 28.33 % after 2 and 4 days. The metal removal capacity per CFU was the maximum for lead and arsenic compared to other metals, which were 1.99E-7 and 1.45E-07. The heat-inactivated bacterial cells removed arsenic in higher concentrations and lead in lower concentrations. The electron microscopy showed no significant alteration in bacterial morphology in control and metal-treated bacterial cells. The nanopore long-read sequencing analysis revealed that cadmium, nickel, copper, and arsenic resistance genes were found on the bacterial genome. No genes were found for lead and chromium but 849 hypothetical coding sequences having unknown functions were present on the bacterial genome. So, the Microbacterium sp. strain 1S1 is a potential candidate for the removal of heavy metals from e-waste dumping sites.

|
Scooped by
?
June 22, 2:45 PM
|
The gut virome plays a pivotal role in shaping the host’s microbiota. However, gut viruses across different mammalian models, and their connections with the human gut microbiota remain largely unknown. We identified 977 high-confidence species-level viral operational taxonomic units (vOTUs) in mice (hcMGV), 12,896 in pigs (hcPGV), and 1480 in cynomolgus macaques (hcCMGV) from metagenomes, respectively. Clustering these vOTUs at approximately genus level uncovered novel clades with high prevalence across animal guts (> = 60%). In particular, crAss-like phages and cas-harboring jumbophages were characterized. Comparative analysis revealed that hcCMGV had a closer relationship with hcPGV than hcMGV, despite the animal-specific characteristics, and that 55.88% hcCMGV had connections with the human microbiota. Our findings shed light on the diversity of gut viruses across these three animals, contributing to future gut microbial studies using model animals.

|
Scooped by
?
June 22, 2:39 PM
|
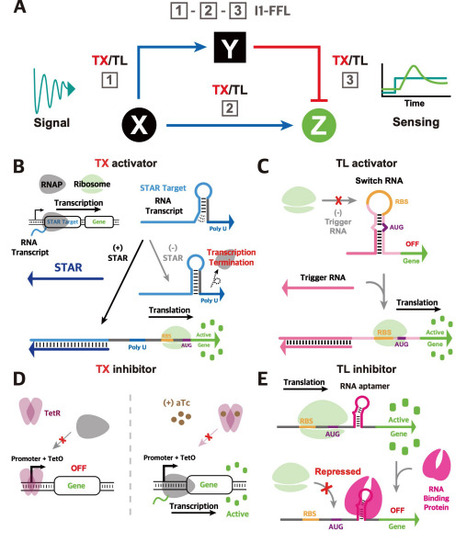
Regulating gene expression with precision is essential for cellular engineering and biosensing applications, where rapid, programmable, and sensitive control is desired. Current approaches to regulatory circuit design often rely on control at a single regulatory level, primarily the transcriptional level, thereby limiting the capability of fine-tuning the regulatory dynamics in response to complex stimuli. To address this challenge, we developed four novel RNA-protein hybrid type-1 incoherent feed-forward loop (I1-FFL) circuits in Escherichia coli that integrate transcriptional and translational regulators to achieve multi-level control of gene expression. These hybrid circuits leverage the modularity and rapid dynamics of RNA-based activators alongside the versatile inhibition capabilities of the protein-based repressors, to endow tunable pulse dynamics through engineered delays that act as transient repressor decoys. By repurposing synthetic RNA regulators at multiple regulatory levels together with aptamer and RNA-binding proteins, we demonstrate previously unexplored circuits with tunable dynamics. Complementary simulation results highlighted the importance of the engineered delays in achieving tunable pulse dynamics in these circuits. Integrating modeling insights with experimental validation, we demonstrated the flexibility of designing the RNA-protein hybrid I1-FFL circuits, as well as the tunability of their dynamics, highlighting their suitability for applications in environmental monitoring, metabolic engineering, and other engineered biological systems, where precise temporal control and adaptable gene regulation are desired.

|
Scooped by
?
June 22, 2:27 PM
|
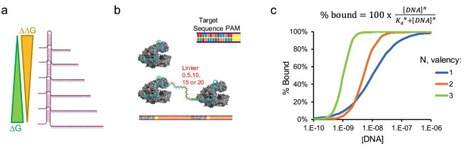
CRISPR-Cas diagnostics are revolutionizing point-of-care molecular testing due to the programmability, simplicity, and sensitivity of Cas systems with trans-cleavage activity. CRISPR-Cas12 assays are promising for detecting single nucleotide polymorphisms (SNPs). However, reports vary widely describing Cas12 SNP sensitivity, and an underlying mechanism is lacking. We systematically varied crRNA length and valency to investigate the role of crRNA architectures on Cas12 biosensing in the context of speed-of-detection, sensitivity, and selectivity. Our results demonstrate that crRNAs complementary to 20 base pairs of the target DNA is optimal for rapid and sensitive detection, while a complementary length of 15 base pairs is ideal for robust SNP detection. Additionally, we uncovered a unique periodicity in SNP sensitivity based on nucleotide position and developed a structural model explaining what drives Cas12 SNP sensitivity. Lastly, we showed that bivalent CRISPR-Cas sensors have synergistic and enhanced activity that is distance dependent. Profiling crRNAs revealed optimal length for Cas12-based molecular testing and SNP detection. Investigation into multivalent architectures elucidated valency and distance dependence for synergistic Cas12 activity.

|
Scooped by
?
June 22, 2:18 PM
|
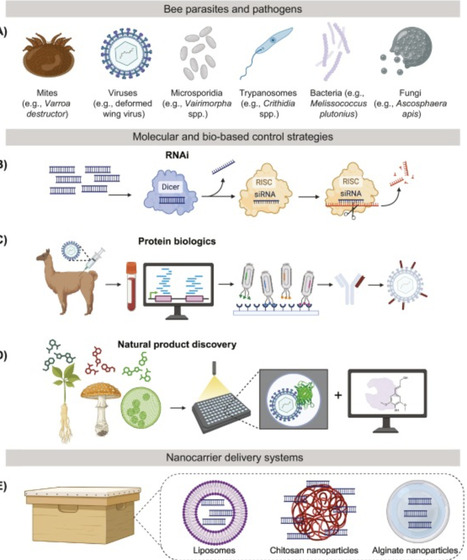
Bees are vital to global food security and biodiversity but their populations are threatened by a steady flux of interacting stressors. Current mitigation strategies are failing to address the complexity and scale of these threats. Biotechnology offers innovative solutions to protect essential pollination services and secure the future of beekeeping. Omic tools guided by artificial intelligence can unlock new possibilities for strengthening bee populations and improve their ability to adapt to emerging challenges. Molecular and bio-based treatments offer precise, nonchemical inputs for managed hives. Synthetic biology enables engineered gut microbiomes, pollinator-friendly crops, and artificial diets that are tailored to bee health. We discuss recent progress and future directions of biotechnology to help bees cope with a rapidly changing world.

|
Scooped by
?
June 22, 1:31 PM
|
Cyanidiophyceae red algae dominate many geothermal habitats and provide important tools for investigating the evolution of extremophilic eukaryotes and associated microbial communities. We propose that resource sharing drove genome reduction in Cyanidiophyceae and enabled the neofunctionalization of genes in multi-enzyme pathways. Utilizing arsenic detoxification as a model, we discuss how the sharing of gene functions by other members of the microbial assemblage weakened selection on homologs in the Cyanidiophyceae, allowing long-term gene persistence via the putative gain of novel functions. This hypothesis, referred to as the Integrated Horizontal Gene Transfer (HGT) Model (IHM), attempts more generally to explain how extremophilic eukaryotes may have transitioned from 'hot start' milieus by functional innovations driven by the duplication and divergence of HGT-derived genes.

|
Scooped by
?
June 22, 1:26 PM
|
Big Science projects are often troubled by misalignments. We analyze how synthetic cell researchers handle misalignments by performing alignment work. We find that alignment work renders Big Science feasible, but involves trade-offs, for example between short-term successes and long-term goals. We thus recommend attending to the politics of alignment work.

|
Scooped by
?
June 22, 1:13 PM
|
Traditionally perceived as an RNA-specific nuclease, Cas13a has been used primarily for RNA detection. We discover the ability of Leptotrichia buccalis Cas13a (LbuCas13a) to directly target DNA without the restrictions of protospacer flanking sequence and PAM sequences, coupled with robust trans-cleavage activity. Contrary to conventional understanding, LbuCas13a does not degrade DNA targets. Our study reveals an enhancement in the single-nucleotide specificity of LbuCas13a against DNA compared to RNA. This heightened specificity is attributed to the lower affinity of CRISPR RNA (crRNA) towards DNA, raising the crRNA–DNA binding energy barrier. We introduce a molecular diagnostic platform called superior universal rapid enhanced specificity test with LbuCas13a (SUREST) for high-resolution genotyping. SUREST is capable of detecting DNA concentrations of CYP2C19 (rs4986893) as minute as 0.3 aM (0.18 cps µl−1). We also apply SUREST to human genotyping scenarios, indicating that SUREST performs well across a broad range of mutations and sequence contexts. SUREST represents an advancement in real-time nucleic acid detection, making it a useful tool for pathogen identification and mutation analysis in clinical diagnostics. A Leptotrichia buccalis Cas13a diagnostic platform can be used to cleave and detect small concentrations of viral and human DNA.
|
 Your new post is loading...
Your new post is loading...