Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 11:14 AM
|
Cas9-based genome engineering is a powerful tool for yeast strain development, but its use in the food industry is limited due to GMO concerns. As a non-GMO alternative, adaptive laboratory evolution (ALE) was applied to enhance trehalose accumulation, a stress protectant, in Saccharomyces cerevisiae. To avoid ethanol-centric metabolism and promote storage carbohydrate production, ALE was conducted under nitrogen limitation with ethanol as the sole carbon source. The evolved strain 65EV showed a 2.3-fold increase in trehalose (11.51% vs 5.25%), resulting in enhanced cell viability (77.7% vs 5.11%) and bread loaf volume (107.2 mL vs 61.5 mL) after freeze/thaw stress. Amino acid profiling revealed distinct metabolic shifts, including elevated intracellular proline and extracellular glutamate. Whole-genome sequencing and reverse engineering identified unique mutations, TSL1 (V887A) and SSA2 (F105L), associated with trehalose regulation. These findings demonstrate potential of ALE as a non-GMO strategy for improving yeast performance in food applications.

|
Scooped by
mhryu@live.com
Today, 11:08 AM
|
Maintaining proper redox conditions is essential for protein stability and function. In cell-free protein synthesis, reducing agents, such as dithiothreitol and reduced glutathione, are commonly added to mimic the cytosolic environment and prevent unwanted oxidation. The PURE system, which is a fully reconstituted protein synthesis system, also contains reducing agents. Here, we systematically examined how reducing agents affect the protein synthesis in the PURE system. We found that the reducing activity of dithiothreitol decreased during prolonged reactions, leading to the formation of disulfide bonds in synthesized proteins. Dissolved oxygen and contaminating metal ions were identified as major factors causing this loss of activity. Based on these findings, we developed a method to maintain reducing conditions throughout the reaction, ensuring consistent protein quality. Our results provide new insights into redox regulation in cell-free systems and offer a practical strategy for the efficient synthesis of functional proteins, with potential applications in biotechnology and therapeutic protein production.

|
Scooped by
mhryu@live.com
Today, 11:00 AM
|
Annotation of genes and transcripts is a key prerequisite for understanding the information that is encoded in newly sequenced genomes. One source of information suited for this purpose is RNA-seq data mapped to the respective genome sequence. RNA-seq-based approaches for transcript reconstruction generate transcript models from these data by combining regions of contiguous coverage (exons) and split read mappings (introns). Understanding phenotypes as a consequence of proteins encoded in a genome further requires the annotation of coding sequences within transcript models. We present GeMoSeq, a novel approach for transcript reconstruction from RNA-seq data that combines combinatorial enumeration of candidate transcripts with heuristics for splitting candidate transcripts into regions of contiguous coverage and subsequent likelihood-based quantification. Prediction of coding sequences is an integral part of the GeMoSeq algorithm. We benchmark GeMoSeq against previous approaches using a large collection of public RNA-seq data for seven species. For the majority of species, we observe an improved prediction performance of GeMoSeq, especially on the level of coding sequences and for species with dense genomes. We combine GeMoSeq with the homology-based approach GeMoMa to re-annotate two recently sequenced genomes of Nicotiana benthamiana lab strains, which illustrates the main purpose of GeMoSeq: the initial annotation of newly sequenced genomes with protein-coding genes.

|
Scooped by
mhryu@live.com
Today, 10:31 AM
|
Extracellular electron transfer (EET) is essential for electroactive microbes’ physiology and biotechnological applications. Many such microbes are Gram-negative bacteria, in which EET must cross two membranes and the periplasm, necessitating spatial and temporal collaborations of various EET proteins that reside at different cellular compartments, for which little is known. Using single-molecule/single-cell-level fluorescence microscopy and electrochemical manipulations, we discover that in the electroactive bacterium Shewanella oneidensis, the inner-membrane electron-transfer hub protein CymA undergoes spatial reorganization into localized regions during active EET with dispersed formation dynamics, subsequently driving the colocalization of its direct electron-transfer partners in the periplasm. Correlated single-cell-level photoelectrochemistry-fluorescence microscopy further proves the critical function of CymA reorganization in enabling EET. A multitude of evidence suggests that CymA reorganization stems from biomolecular condensate formation, likely initiated by association with menaquinone-rich inner-membrane domains. These orchestrated spatiotemporal protein dynamics extend the functional roles of biomolecular condensates to include facilitation of EET in bacteria, with broader implications for cellular processes. Extracellular electron transfer (EET) in bacteria requires the spatiotemporal coordination of many proteins. Here, authors show that the inner-membrane protein CymA reorganizes spatially into condensates and drives the colocalization of partner proteins to enable EET in Shewanella.

|
Scooped by
mhryu@live.com
Today, 10:07 AM
|
Genome annotation captures the essence of a genome by cataloguing its genes, transcripts, proteins and other functional elements of the DNA sequence. Accurate annotation serves as the foundation for a wide range of downstream analyses and discoveries, ranging from basic biology to an understanding of the linkage between genes and disease. Over the past two decades, advances in high-throughput sequencing techniques have enabled faster and more accurate capture of diverse genomic features, generating data at an unprecedented scale. Concurrently, computational methods for translating these data into evidence for genome annotation have steadily improved, leading to better automated genome annotation systems. As such, the growing number of sequenced genomes provides a positive feedback loop, in which database searches become more effective and shared sequence patterns emerge more clearly. These advances are promising steps towards annotating the functions of many poorly understood genes, particularly non-coding RNA genes, for which more research is needed. In this Review, Ji et al. overview how rapidly advancing experimental and computational methods are enabling improved and automated annotation of gene structure and function, providing researchers with genome annotation resources of unprecedented scale and resolution.

|
Scooped by
mhryu@live.com
Today, 9:34 AM
|
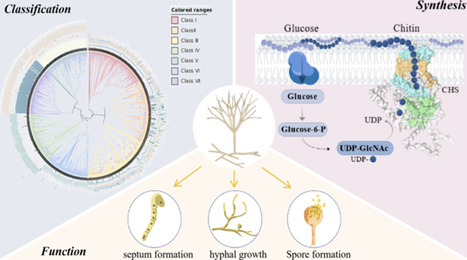
Fungi contain the most complex chitin synthase family, which contributes to chitin biosynthesis and cell wall rigidity. In the past decades, their functions involved in morphological diversity, viability, and virulence of fungi have been well investigated using typical molecular methods such as gene knockout or gene silence, which seems to give a simple aspect of their comprehensive roles. The present review summarized the comprehensive advances of chitin synthases including their classification, their precise structures including catalytic and noncatalytic protein domains, the catalytic mechanisms of chitin biosynthesis and translocation, and roles in cell wall formation, cell separation, and cell viability to provide the references for understanding and guiding the discovery of the novel antifungal drugs targeting the chitin synthases according to their structural basis to offer significant effects and lower resistance development.

|
Scooped by
mhryu@live.com
Today, 1:23 AM
|
Here, we introduce Detectrons, modular biosensors that couples programmable toehold switches with retron-mediated reverse transcription to transduce RNA inputs into unique DNA barcodes. The ability to convert dynamic RNA signals into durable DNA records within living cells unlocks powerful new modes of transcript-based sensing with applications including viral infection detection. Through the construction of a synthetic toehold-retron library and application of machine learning, we uncovered key design principles that improve signal strength and specificity. We applied Detectrons to the multiplexed live-cell detection of specific phage infections, enabling transcript-triggered barcode synthesis and quantitative host susceptibility profiling in pooled bacterial populations. Detectrons are the first RNA-to-DNA transduction system, directly linking transient RNA detection to stable, sequence-encoded DNA outputs. This platform provides a scalable and generalizable strategy for phage screening and for recording transcriptional events in complex bacterial communities.

|
Scooped by
mhryu@live.com
Today, 1:15 AM
|
Conventional plant genome editing relies on tissue culture-mediated somatic cell regeneration, a technically demanding process that limits its application across diverse species. Emerging strategies now circumvent this bottleneck by enabling direct genome editing of meristematic or germline cells. Key advances include (i) genome editing via de novo meristem induction or dormant meristem activation; (ii) germline editing facilitated by graft-mobile tRNA-like sequence systems and haploid induction technologies; and (iii) optimized viral delivery platforms that exploit mobile RNA elements and compact editors such as TnpB to achieve efficient, transgene-free, heritable modifications across a broad range of genotypes and species. The development of robust, tissue culture-free editing platforms promises to revolutionize crop improvement pipelines and accelerate trait development for sustainable agriculture.

|
Scooped by
mhryu@live.com
Today, 1:06 AM
|
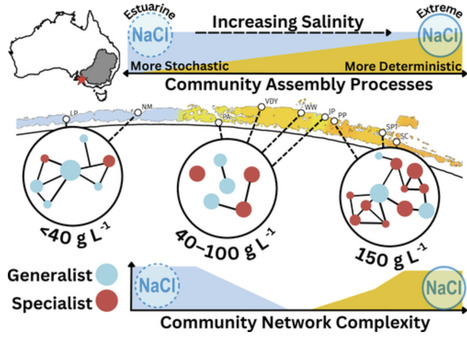
Coastal wetlands are highly vulnerable to climate-driven salinization, which reshapes critical microbial processes underpinning nutrient cycling and energy flow. We examined how sediment microbial communities vary with salinity across the Coorong Lagoon (South Australia), spanning estuarine (0–40 g L−1), intermediate (40–100 g L−1) and hypersaline (100–150 g L−1) waters. Salinity was found to be the dominant driver of sediment microbial community composition, diversity and assembly. High salinity favored specialists and homogenous community structures, with generalist bacteria persisting across intermediate salinities and supporting ecosystem resilience. Sulfur and carbon cycling is likely dependent on salinity, as bacterial sulfur-oxidizers were abundant estuarine specialists, whereas methane producers (Archaeal methanogens) and sulfate-reducers were enriched at high salinity. Deterministic microbial community assembly (homogeneous selection) was dominant, increasing at extreme salinity, which acted as a strong environmental filter. Community complexity increased at both high and low salinity ranges, with intermediate salinity exhibiting less complexity, suggesting community reorganisation under osmotic stress. The varied roles of specialists and generalists at different salinities support ecosystem function, where increased heterogeneity and specialization in hypersaline conditions suggest vulnerability of the community to disturbance. These findings provide insight into how microbially underpinned ecosystems may respond to future climate-driven salinization, important for making predictions and informing mitigation strategies.

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
The ability to engineer synthetic biomolecular condensates in living cells offers new opportunities to control intracellular organization, yet robust and programmable RNA-based systems have remained limited. Here, we introduce genetically encoded, modular platforms that generate RNA-driven condensates using nanostar-derived scaffolds. Systematic comparison of repeat-based and de novo designs identified nanostar variants that reliably assemble nuclear condensates in mammalian cells. Unexpectedly, condensate formation in cells is governed primarily by double-stranded RNA stems that recruit endogenous RNA-binding proteins, rather than by the kissing-loop interactions that drive assembly in vitro. This mechanistic shift highlights the divergence between cellular and in vitro environments and accounts for the limited orthogonality among scaffolds. Sequence refinement to reduce nonspecific pairing improved homotypic assembly and enhanced orthogonality. We further demonstrated functional compartmentalization by recruiting protein and RNA clients to modulate their stability and activity, and we incorporated an acyclovir-responsive allosteric switch to achieve reversible, small-molecule control of condensation. Together, this work establishes a versatile RNA-based toolkit for constructing programmable cellular compartments, advancing strategies for controlling RNA-protein organization and enabling new biosensing and therapeutic applications.

|
Scooped by
mhryu@live.com
Today, 12:40 AM
|
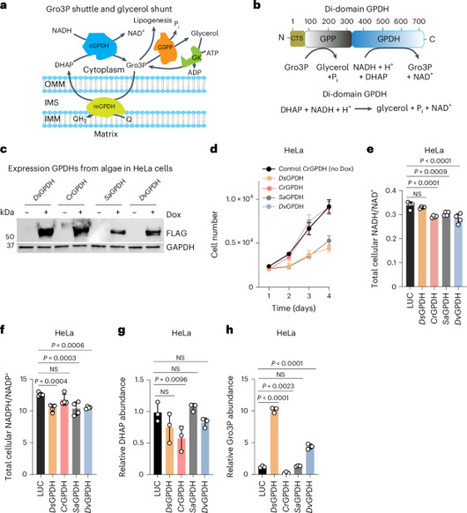
Dihydroxyacetone phosphate (DHAP), glycerol-3-phosphate (Gro3P) and reduced/oxidized nicotinamide adenine dinucleotide (NADH/NAD⁺) are key metabolites of the Gro3P shuttle, which transfers reducing equivalents between the cytosol and mitochondria. Targeted activation of Gro3P biosynthesis has recently emerged as a promising strategy to alleviate reductive stress. However, because Gro3P constitutes the backbone of triglycerides, its accumulation can promote extensive lipogenesis. Here we show that a genetically encoded tool based on a di-domain glycerol-3-phosphate dehydrogenase from the alga Chlamydomonas reinhardtii (CrGPDH) effectively operates both the alternative Gro3P shunt, which regenerates NAD⁺ while converting DHAP to Gro3P, and the glycerol shunt, which converts Gro3P to glycerol and inorganic phosphate, across transformed and primary mammalian cell cultures as well as mouse liver. CrGPDH expression supported proliferation of cancer cells under respiratory chain inhibition or hypoxia, as well as patient-derived fibroblasts with mitochondrial dysfunction. Moreover, CrGPDH decreased triglyceride levels in kidney cancer cell lines and reversed ethanol-induced triglyceride accumulation in mouse liver. Thus, CrGPDH represents a promising xenotopic tool to alleviate redox imbalance and associated impaired lipogenesis in conditions ranging from primary mitochondrial diseases to steatosis. The authors present a genetically encoded tool based on a bifunctional enzyme that can regenerate NAD+ while executing an engineered glycerol shunt. The tool successfully restored redox imbalance and modulated lipid metabolism in vitro and in a mouse hepatic steatosis model.

|
Scooped by
mhryu@live.com
Today, 12:31 AM
|
The demand for rapid and scalable biosensing technologies has motivated the development of antibody-free platforms capable of operating in complex sample environments. Here, we report an electrochemical biosensor based on engineered M13 bacteriophages displaying a SARS-CoV-2 spike S1–binding peptide immobilized on a reduced graphene oxide (rGO) transducer. The sensor employs a chemiresistive detection mechanism under a fixed low-voltage bias, enabling rapid electrical readout following target binding. Detection of S1 protein was achieved in buffer and in spiked complex matrices, including fetal bovine serum, pasteurized milk, and wastewater, demonstrating matrix tolerance under the tested conditions. The biosensor response is evaluated using a statistically defined binary detection criterion, with an operational limit of detection of 10⁻4 pg/mL in buffer. Compared to a previously reported antibody-functionalized rGO sensor fabricated using the same platform, the phage-based biosensor exhibits comparable sensitivity while offering advantages in genetic tunability and production scalability. While the present study focuses on proof-of-concept validation using spiked samples, these results highlight the potential of engineered phage–graphene interfaces as adaptable biorecognition elements for rapid electrochemical protein sensing in complex environments.

|
Scooped by
mhryu@live.com
February 16, 10:59 PM
|
Achieving neutral nitrogen (N) budgets is critical for soil health and sustainable agriculture. Thus, 31 studies were conducted for soybeans (Glycine max L.) throughout the United States, determining N inputs, biological N2 fixation via δ15N natural abundance method, and N outputs, seed N removal. On average, 53% of the N demand was met through N2 fixation, while the requirement of 62% indicates a significant shortfall for achieving neutral N budgets.
|

|
Scooped by
mhryu@live.com
Today, 11:11 AM
|
Lactic acid bacteria (LAB), a group of Gram-positive bacteria widely used in food fermentation, are major producers of bacteriocins─ribosomally synthesized antimicrobial peptides. These peptides have attracted considerable attention not only as natural food preservatives but also for their therapeutic potential in biomedicine. This review outlines LAB-derived bacteriocins, presenting an updated classification (Classes I–III) by structure and genetics, along with their biosynthesis and gene clusters. Furthermore, key production influencers, such as quorum sensing, two-component systems, and culture conditions, are analyzed. Besides, this review also highlights the health benefits of LAB bacteriocins against infections and cancers, achieved via mechanisms like membrane disruption. It also discusses strategies (e.g., metabolic engineering and synthetic biology, combination therapies, and novel purification methods) to enhance efficacy. Despite persistent challenges in yield optimization and clinical translation, this work provides critical insights to guide the therapeutic development and scalable production of bacteriocins.

|
Scooped by
mhryu@live.com
Today, 11:05 AM
|
Non-genetic variability in gene expression is an inevitable consequence of the stochastic nature of processes driving transcription and translation. While previous studies demonstrated that gene expression noise is negatively correlated with gene body methylation, the function of this correlation remains poorly understood in multicellular systems. Here, we provide a first functional link between gene body methylation and transcription noise in plants. We investigated a mutant with partial loss of CG methylation (met1-1) and 10 epigenetic recombinant inbred lines (epiRILs) generated by a cross between Col-0 and met1-3 plants, and observed an increase in gene expression noise, but this was not the case in met1-3 with complete loss of CG methylation. Loss of CG methylation in met1-3 could be compensated by a low but significant gain of non-CG methylation that buffers the noise in gene expression. Overall, our results show that gene body methylation has a functional role in reducing variability in transcription in a large subset of housekeeping genes, which require precise expression patterns to meet metabolic requirements. Genes lacking this noise-buffering effect are mainly enriched in stress response, where variability in gene expression can be seen as highly beneficial.

|
Scooped by
mhryu@live.com
Today, 10:36 AM
|
Unconventional protein secretion (UcPS) enables the export of cytosolic proteins through pathways that bypass the canonical endoplasmic reticulum–Golgi secretory route. Although increasingly recognized as essential for intercellular communication, stress responses, and tissue homeostasis, UcPS remains difficult to quantify due to low secretion efficiency, high intracellular background, and the challenge of distinguishing active secretion from passive leakage. Recent methodological advances, including NanoLuc split luciferase–based reporters and the Retention Using Selective Hooks (RUSH) system for synchronized protein transport, have improved sensitivity and temporal control of trafficking. Here, we present complementary protocols integrating these tools to provide a highly sensitive, quantitative workflow centered on a split NanoLuc (HiBiT/LgBiT) complementation assay for monitoring UcPS in mammalian cells. The Basic Protocol describes a robust luminescence-based secretion assay, while the Support Protocols detail the generation of stable HiBiT reporter cell lines, approaches for probing UcPS mechanisms using siRNA-mediated gene knockdown and pharmacological perturbation, and the incorporation of the RUSH system to synchronize cargo release and identify potential trafficking intermediates. Together, these protocols provide a sensitive, scalable, high-throughput toolkit that enables analysis of UcPS mechanisms across diverse cargo proteins, cell types, and perturbations. This methodological framework allows for rigorous dissection of UcPS pathways in both physiological and disease-relevant contexts.

|
Scooped by
mhryu@live.com
Today, 10:28 AM
|
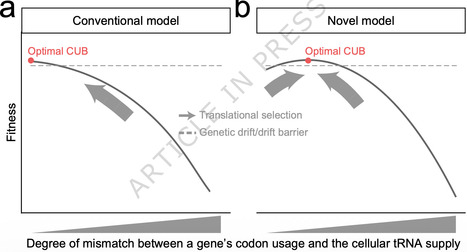
Each amino acid except two is encoded by multiple synonymous codons, but at unequal frequencies. Such codon usage bias (CUB) is observable in almost all species, and commonly assumed as the result of natural selection towards an optimal CUB that matches the cellular tRNA supply. Here we hypothesize instead that the optimal CUB of a gene should slightly mismatch the tRNA supply to avoid excessive translational costs, while ensuring adequate functional payoff. By modifying the CUB of a resistance gene expressed in bacteria under antibiotic selection, we demonstrate that a small mismatch with the tRNA supply confers faster bacterial growth than those with minimized or large CUB-tRNA mismatches. Intriguingly, the optimal degree of CUB-tRNA mismatch increases as the resistance gene becomes less important in media with lower antibiotic concentrations, which is explainable by our model as a shift in the balance between the gene’s functional payoff and translational cost. Furthermore, genomic analyses in model organisms suggest that the optimal degree of CUB-tRNA mismatch is larger for endogenous genes with lower functional importance and higher mRNA abundance, respectively supporting the impact of functional payoff and translational cost. Finally, we find that mutations increasing or decreasing the CUB-tRNA mismatch of native genes are both predominantly deleterious, such that the CUB-tRNA mismatch is likely selectively maintained rather than minimized to that achievable in the presence of genetic drift and mutational bias. These results challenge the commonly assumed unidirectional selection on CUB and highlight the CUB-modulated balance between functional payoff and translational cost. Nearly all organisms exhibit codon usage bias (CUB), traditionally thought to reflect a unidirectional selection maximally matching CUB and the cellular tRNA supply. Here, the authors propose alternatively that a slight mismatch optimizes the balance between translational cost and functional payoff.

|
Scooped by
mhryu@live.com
Today, 9:36 AM
|
3-Hydroxy-3-methylglutaryl-CoA synthase (HMGS) is a key enzyme in the mevalonate (MVA) pathway that catalyzes the formation of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) from acetoacetyl-CoA and acetyl-CoA. Recently, a novel class of HMGS-sesquiterpene synthase (STS) fusion enzymes has been identified. In this study, we discovered a natural fusion enzyme, GihirA, in Gloeostereum incarnatum, which contains both STS and HMGS domains and synthesizes the sesquiterpenoid hirsutene. Our investigation revealed that the HMGS domain significantly enhances the cyclization activity of the STS domain, resulting in an 8.87-fold increase in sesquiterpene production with a final yield of 121.3 mg/kg, highlighting HMGS’s critical role in catalytic efficiency. Additionally, domain-swapping experiments were performed by replacing the HMGS domain of G. incarnatum with the native HMGS domain from Flammulina velutipes sesquiterpene synthase Fla2. The results demonstrated that Fla2 fused with its cognate HMGS domain exhibited a significant yield enhancement from 11.5 to 54.5 mg/kg, underscoring the importance of metabolic compatibility in enzyme performance. This study not only reveals the unique advantages of natural fusion enzymes in sesquiterpene biosynthesis but also provides an important theoretical foundation for enhancing sesquiterpene production through the optimization of enzyme fusion strategies and metabolic pathway design. These findings offer a rational strategy for engineering high-efficiency terpenoid biosynthesis.

|
Scooped by
mhryu@live.com
Today, 1:31 AM
|
Gene duplication is the primary mechanism by which new genes emerge. Models and empirical studies have shown that paralogous genes are maintained because of dosage benefits, the partitioning of ancestral functions or the acquisition of new functions. However, the underlying molecular mechanisms and the relative importance of the factors driving evolution towards one fate or another have remained difficult to quantify. Recent advances in experimental and computational methods, such as gene editing, deep mutational scanning and ancestral sequence reconstruction, have enabled molecular analyses of duplicated gene evolution across timescales. Combined, these approaches are revealing how adaptive and non-adaptive evolutionary forces shape the modern fates of gene duplicates. Gene duplication is a key evolutionary mechanism, as initially redundant paralogues diverge over time. The authors review how adaptive and non-adaptive forces influence the evolutionary fates of gene duplicates, highlighting the importance of function–fitness relationships and gene expression dynamics.

|
Scooped by
mhryu@live.com
Today, 1:20 AM
|
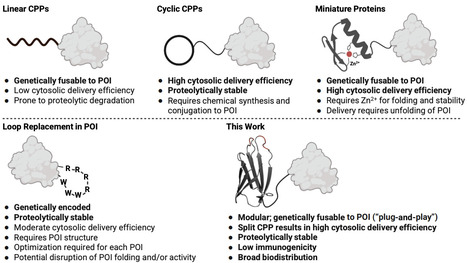
Antibodies and other protein therapeutics have revolutionized medicine, but their application is largely limited to extracellular targets. The lack of efficient intracellular delivery methods remains a major bottleneck. Here, we engineered a family of small (~90 amino acids), metabolically stable membrane translocation domains (MTDs) by modifying the loop sequences of a human fibronectin type III (FN3) domain. The most potent variant, MTD4, is highly cell-permeable and can be recombinantly fused to the N- or C-terminus of any peptide or protein, serving as a versatile "plug-and-play" vehicle. We demonstrate that MTD4 fusions efficiently deliver a wide variety of functional peptides and proteins into the cytosol and nucleus of eukaryotic cells, both in vitro and in vivo. Following systemic administration, MTD4 fusion proteins exhibit broad biodistribution and homogenous tissue penetration in mice. Importantly, MTD4 is effective at low nanomolar (nM) concentrations, making it a promising platform for addressing a vast range of intracellular and previously "undruggable" targets.

|
Scooped by
mhryu@live.com
Today, 1:10 AM
|
5-Aminolevulinic acid (5-ALA), the universal precursor of tetrapyrroles, has widely applications such as photodynamic therapy of skin cancer, agriculture booster and cosmetics. 5-ALA production relies on the efficiency of 5-aminolevulinate synthase (ALAS) and energy balance for cell growth. In this study, the folding capacity of ALAS was enhanced by chromosomal integration of molecular chaperone systems, including Trigger Factor (TF), GroEL/GroES (GroELS), and DnaK/DnaJ (DnaKJ), in engineered E. coli expressing Rhodobacter capsulatus ALAS (RcALAS). Among all systems, the DnaKJ-integrated strain markedly improved solubility, further plasmid-based co-expression of DnaKJ increased 5-ALA production to 7.15 g/L, representing a 2.23-fold increase over the control. By refining carbon source utilization and feeding strategies to encounter the ATP demand, 5-ALA titer was further increased to 9.36 g/L within 36 h in shake flask cultivation. To validate the process performance, bench-scale cultivation in a 2.5 L Ultra-Yield flask demonstrated its scalability, achieving a final 5-ALA titer of 12.1 g/L. The successful 5-ALA biosynthesis assisted by DnaKJ chaperone, coupled with carbon flux redirection, showed a synergistic effect that improved heterologous enzyme expression and evaluated overall efficiency of 5-ALA production.

|
Scooped by
mhryu@live.com
Today, 1:04 AM
|
Pseudomonas, a cornerstone genus in petroleum hydrocarbon bioremediation, exhibits remarkable metabolic diversity. To systematically decipher the genetic basis of this trait, we constructed a curated collection of representative Pseudomonas strains with documented degradation capabilities through a bibliometrics-driven approach. Comparative genomic analysis revealed that these strains possess a rich repertoire of genes linked to petroleum hydrocarbon degradation, including those encoding key enzymes such as monooxygenases, dioxygenases, alcohol dehydrogenases, cytochrome P450, ferredoxins, and regulatory proteins (e.g., LuxR, AraC, GntR). Among the strains examined, P. citronellolis and P. putida contained the highest abundance of such genes. The accessory genome size varied considerably across the 15 strains (ranging from 3290 to 5745 genes), and functional enrichment analysis indicated a significant concentration of degradation-related genes within this component. This genomic architecture not only reflects distinct metabolic specializations among species but also implies potential synergistic interactions, as suggested by the broader genetic accessibility to polycyclic aromatic hydrocarbon (PAH) degradation pathways observed in P. aeruginosa, P. luteola, and P. putida. Overall, this study establishes a robust genomic framework that extends beyond single-species analysis, offering a genus-level perspective essential for designing tailored, high-efficiency microbial consortia for targeted bioremediation strategies.

|
Scooped by
mhryu@live.com
Today, 12:47 AM
|
Global freshwater scarcity and degradation demand reconceptualizing wastewater as a resource within circular economy transitions. This review utilizes the resource-stack model and treatment-train design space to evaluate wastewater valorization, assessing recovery of water, energy, nutrients, carbon, and trace elements. It examines barriers, inefficiencies, costs, regulatory gaps, and societal resistance and highlights that aligning innovation with policy and governance can transform wastewater systems into infrastructures supporting water security and climate resilience.

|
Scooped by
mhryu@live.com
Today, 12:34 AM
|
Cellular immunotherapies encompass a broad and rapidly developing group of treatments comprising expanded and/or genetically engineered immune cells, which use the specific properties of human immune cells to counteract human immune-mediated disease. Initially approved for cancers of the B cell lineage, a growing arsenal of cellular immunotherapies are being applied to autoimmune diseases, including chimeric antigen receptor (CAR) T cells, chimeric autoantibody receptor T cells, regulatory T cells and CAR-engineered innate immune cells. These approaches represent a major shift in the way scientists and physicians pursue the treatment of human disease compared to standard immunosuppressive therapies. Here, we review the clinical progress of engineered cellular immunotherapies for autoimmunity. We focus on how antigenic target, engineered cell type, CAR design and treatment regimens affect the therapeutic efficacy and safety of these treatments and how these emerging clinical data can inform future directions in the field. Engineered cellular immunotherapy shows great potential for treating autoimmune diseases. Payne and colleagues review emerging clinical data of the most recently developed technologies.

|
Scooped by
mhryu@live.com
February 16, 11:05 PM
|
Klebsiella pneumoniae intestinal colonization contributes to infectious and non-infectious diseases. Recent studies have uncovered the complex interplay between the gut microbiota and K. pneumoniae colonization, highlighting its role in disease development and progression and illustrating the potential of microbiome-based therapies. Yang et al. summarize that Klebsiella pneumoniae affects infectious and non-infectious disease via gut colonization; commensals provide colonization resistance, but K. pneumoniae adapts via multiple mechanisms.
|
 Your new post is loading...
Your new post is loading...