 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:31 AM
|
Gene duplication is the primary mechanism by which new genes emerge. Models and empirical studies have shown that paralogous genes are maintained because of dosage benefits, the partitioning of ancestral functions or the acquisition of new functions. However, the underlying molecular mechanisms and the relative importance of the factors driving evolution towards one fate or another have remained difficult to quantify. Recent advances in experimental and computational methods, such as gene editing, deep mutational scanning and ancestral sequence reconstruction, have enabled molecular analyses of duplicated gene evolution across timescales. Combined, these approaches are revealing how adaptive and non-adaptive evolutionary forces shape the modern fates of gene duplicates. Gene duplication is a key evolutionary mechanism, as initially redundant paralogues diverge over time. The authors review how adaptive and non-adaptive forces influence the evolutionary fates of gene duplicates, highlighting the importance of function–fitness relationships and gene expression dynamics.

|
Scooped by
mhryu@live.com
Today, 1:20 AM
|
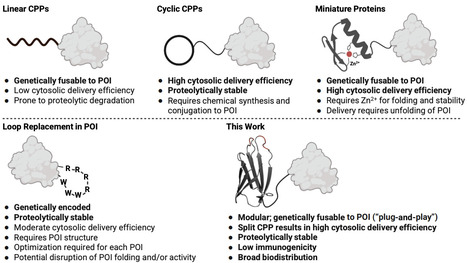
Antibodies and other protein therapeutics have revolutionized medicine, but their application is largely limited to extracellular targets. The lack of efficient intracellular delivery methods remains a major bottleneck. Here, we engineered a family of small (~90 amino acids), metabolically stable membrane translocation domains (MTDs) by modifying the loop sequences of a human fibronectin type III (FN3) domain. The most potent variant, MTD4, is highly cell-permeable and can be recombinantly fused to the N- or C-terminus of any peptide or protein, serving as a versatile "plug-and-play" vehicle. We demonstrate that MTD4 fusions efficiently deliver a wide variety of functional peptides and proteins into the cytosol and nucleus of eukaryotic cells, both in vitro and in vivo. Following systemic administration, MTD4 fusion proteins exhibit broad biodistribution and homogenous tissue penetration in mice. Importantly, MTD4 is effective at low nanomolar (nM) concentrations, making it a promising platform for addressing a vast range of intracellular and previously "undruggable" targets.

|
Scooped by
mhryu@live.com
Today, 1:10 AM
|
5-Aminolevulinic acid (5-ALA), the universal precursor of tetrapyrroles, has widely applications such as photodynamic therapy of skin cancer, agriculture booster and cosmetics. 5-ALA production relies on the efficiency of 5-aminolevulinate synthase (ALAS) and energy balance for cell growth. In this study, the folding capacity of ALAS was enhanced by chromosomal integration of molecular chaperone systems, including Trigger Factor (TF), GroEL/GroES (GroELS), and DnaK/DnaJ (DnaKJ), in engineered E. coli expressing Rhodobacter capsulatus ALAS (RcALAS). Among all systems, the DnaKJ-integrated strain markedly improved solubility, further plasmid-based co-expression of DnaKJ increased 5-ALA production to 7.15 g/L, representing a 2.23-fold increase over the control. By refining carbon source utilization and feeding strategies to encounter the ATP demand, 5-ALA titer was further increased to 9.36 g/L within 36 h in shake flask cultivation. To validate the process performance, bench-scale cultivation in a 2.5 L Ultra-Yield flask demonstrated its scalability, achieving a final 5-ALA titer of 12.1 g/L. The successful 5-ALA biosynthesis assisted by DnaKJ chaperone, coupled with carbon flux redirection, showed a synergistic effect that improved heterologous enzyme expression and evaluated overall efficiency of 5-ALA production.

|
Scooped by
mhryu@live.com
Today, 1:04 AM
|
Pseudomonas, a cornerstone genus in petroleum hydrocarbon bioremediation, exhibits remarkable metabolic diversity. To systematically decipher the genetic basis of this trait, we constructed a curated collection of representative Pseudomonas strains with documented degradation capabilities through a bibliometrics-driven approach. Comparative genomic analysis revealed that these strains possess a rich repertoire of genes linked to petroleum hydrocarbon degradation, including those encoding key enzymes such as monooxygenases, dioxygenases, alcohol dehydrogenases, cytochrome P450, ferredoxins, and regulatory proteins (e.g., LuxR, AraC, GntR). Among the strains examined, P. citronellolis and P. putida contained the highest abundance of such genes. The accessory genome size varied considerably across the 15 strains (ranging from 3290 to 5745 genes), and functional enrichment analysis indicated a significant concentration of degradation-related genes within this component. This genomic architecture not only reflects distinct metabolic specializations among species but also implies potential synergistic interactions, as suggested by the broader genetic accessibility to polycyclic aromatic hydrocarbon (PAH) degradation pathways observed in P. aeruginosa, P. luteola, and P. putida. Overall, this study establishes a robust genomic framework that extends beyond single-species analysis, offering a genus-level perspective essential for designing tailored, high-efficiency microbial consortia for targeted bioremediation strategies.

|
Scooped by
mhryu@live.com
Today, 12:47 AM
|
Global freshwater scarcity and degradation demand reconceptualizing wastewater as a resource within circular economy transitions. This review utilizes the resource-stack model and treatment-train design space to evaluate wastewater valorization, assessing recovery of water, energy, nutrients, carbon, and trace elements. It examines barriers, inefficiencies, costs, regulatory gaps, and societal resistance and highlights that aligning innovation with policy and governance can transform wastewater systems into infrastructures supporting water security and climate resilience.

|
Scooped by
mhryu@live.com
Today, 12:34 AM
|
Cellular immunotherapies encompass a broad and rapidly developing group of treatments comprising expanded and/or genetically engineered immune cells, which use the specific properties of human immune cells to counteract human immune-mediated disease. Initially approved for cancers of the B cell lineage, a growing arsenal of cellular immunotherapies are being applied to autoimmune diseases, including chimeric antigen receptor (CAR) T cells, chimeric autoantibody receptor T cells, regulatory T cells and CAR-engineered innate immune cells. These approaches represent a major shift in the way scientists and physicians pursue the treatment of human disease compared to standard immunosuppressive therapies. Here, we review the clinical progress of engineered cellular immunotherapies for autoimmunity. We focus on how antigenic target, engineered cell type, CAR design and treatment regimens affect the therapeutic efficacy and safety of these treatments and how these emerging clinical data can inform future directions in the field. Engineered cellular immunotherapy shows great potential for treating autoimmune diseases. Payne and colleagues review emerging clinical data of the most recently developed technologies.

|
Scooped by
mhryu@live.com
February 16, 11:05 PM
|
Klebsiella pneumoniae intestinal colonization contributes to infectious and non-infectious diseases. Recent studies have uncovered the complex interplay between the gut microbiota and K. pneumoniae colonization, highlighting its role in disease development and progression and illustrating the potential of microbiome-based therapies. Yang et al. summarize that Klebsiella pneumoniae affects infectious and non-infectious disease via gut colonization; commensals provide colonization resistance, but K. pneumoniae adapts via multiple mechanisms.

|
Scooped by
mhryu@live.com
February 16, 10:43 PM
|
Chemical inducers of proximity (CIPs) stabilize biomolecular interactions, often causing an emergent rewiring of cellular biochemistry. While the discovery of heterobifunctional CIPs is expedited by rational design strategies, molecular glues have relied predominantly on serendipity. We hypothesized that preexisting ligands could be systematically decorated with chemical modifications to discover compounds that recruit proteins to a composite protein–ligand interface. Using sulfur(VI) fluoride exchange-based high-throughput chemistry (HTC) to install 3,163 structurally diverse building blocks onto ENL (eleven-nineteen leukemia) and BRD4 (bromodomain-containing protein 4) ligands, we screened each analog for degrader activity. This revealed dHTC1, an ENL degrader that recruits CRL4CRBN complex through an extended interface of protein–protein contacts and only engages CRBN after pre-forming the ENL:dHTC1 complex. We also identified dHTC3, a molecular glue that selectively dimerizes BRD4 bromodomain 1 to SCFFBXO3, an E3 ligase not previously accessible for chemical rewiring. Altogether, this study introduces HTC as a facile tool to discover new CIPs and new effectors for proximity pharmacology. Molecular glue degraders have consistently been discovered retrospectively, despite their increasing importance. Herein, a high-throughput approach is described that modifies existing ligands into molecular glue degraders.

|
Scooped by
mhryu@live.com
February 16, 6:00 PM
|
Deep learning has successfully been applied to design cis-regulatory elements (CREs) for a few species, but a broadly applicable platform for generating functional promoters for thousands of prokaryotes remains lacking. In this study, we introduce a language model for prokaryotic CREs, referred to as PromoGen2, to design CREs without prior experimental data. PromoGen2 was pretrained on CREs derived from 17 000 prokaryotic genomes. It achieved the highest zero-shot prediction correlation of promoter strength across species, improving the average Spearman correlation from 0.27 to 0.50 compared to the best baseline, while reducing the number of parameters by 103. Artificial CREs designed with PromoGen2 demonstrated a 100% success rate in E. coli, Bacillus subtilis, Bacillus licheniformis, and Agrobacterium tumefaciens. Based on PromoGen2, we developed the Promoter-Factory framework to design promoters from unannotated genomes. Experimental validation showed that most of the promoters designed for Jejubacter sp. L23, a newly isolated halophilic bacterium with no available CREs, were active and capable of driving lycopene overproduction. Additionally, we introduced PromoGen2-proka, a taxonomy-aware model for CRE design based on prokaryotic genera. Experimental validation confirmed its reliable success rate. The combined use of PromoGen2-proka and Promoter-Factory offers a broadly applicable tool for designing CREs for prokaryotes, fulfilling the needs of synthetic biology and microbiology research.

|
Scooped by
mhryu@live.com
February 16, 5:17 PM
|
During CRISPR-Cas adaptation, prokaryotic cells become immunized by the insertion of foreign DNA fragments, termed spacers, into the host genome to serve as templates for RNA-guided immunity. Spacer acquisition relies on the Cas1–Cas2 integrase and accessory proteins, which select DNA sequences flanked by the protospacer adjacent motif (PAM) and insert them into the CRISPR array. It has been shown that in type II-A systems, selection of PAM-proximal prespacers is mediated by the effector nuclease Cas9, which forms a “supercomplex” with the Cas1–Cas2 integrase and the Csn2 protein. Here, we present cryo-electron microscopy structures of the Streptococcus thermophilus type II-A prespacer selection supercomplex in the DNA-scanning and two distinct PAM-bound configurations, providing insights into the mechanism of Cas9-mediated prespacer selection in type II-A CRISPR-Cas systems. Repurposing Cas9 by the CRISPR adaptation machinery for prespacer selection, as characterized here, demonstrates Cas9 plasticity and expands our knowledge of Cas9 biology.

|
Scooped by
mhryu@live.com
February 16, 4:58 PM
|
Intrinsically disordered regions (IDRs) navigate transcription factors (TFs) to their binding sites in genomes, raising the question of how IDR sequences can encode for specific genome recognition. To define the principles of IDR-directed binding, we designed de novo IDRs and tested their activity in directing selective binding across the budding yeast genome. Our de novo IDRs were designed by dispersing hydrophobic amino acids within hundreds of hydrophilic residues, as we found to be required in native TF-directing IDRs. Although showing no alignment-based similarity to native TFs, the de novo IDRs were active in directing genome binding toward a tunable range of targets, as revealed by systematically varying the hydrophobic spread or disorder scaffold. Overall, the 185 synthetic IDRs tested displayed a continuum of sequence-directed binding preferences across hundreds of promoters. Our results open new doors for understanding and engineering selective binding across genomes.

|
Scooped by
mhryu@live.com
February 16, 3:38 PM
|
Therapeutic modalities to programmably increase protein production are in critical need to address diseases caused by deficient gene expression via haploinsufficiency. Restoring physiological protein levels by increasing translation of their cognate messenger RNA would be an advantageous approach to correct gene expression but has not been evaluated in an in vivo disease model. Here, we investigated whether a translational activator could improve phenotype in a Dravet syndrome mouse model, a severe developmental and epileptic encephalopathy caused by SCN1a haploinsufficiency, by increasing translation of the SCN1a mRNA. We identify and engineer human proteins capable of increasing mRNA translation using the CRISPR–Cas-inspired RNA-targeting system (CIRTS) platform to enable programmable, guide RNA-directed translational activation with entirely engineered human proteins. We identify a compact (601 amino acid) CIRTS translational activator (CIRTS-4GT3) that can drive targeted, sustained translation increases up to 100% from three endogenous transcripts relevant to epilepsy and neurodevelopmental disorders. AAV-delivery of CIRTS-4GT3 targeting SCN1a mRNA to a Dravet syndrome mouse model led to increased SCN1a translation and improved survivability and seizure threshold—key phenotypic indicators of Dravet syndrome. This work validates a strategy to address SCN1a haploinsufficiency and emphasizes the preclinical potential of targeted translational activation to address neurological haploinsufficiency.

|
Scooped by
mhryu@live.com
February 16, 12:45 PM
|
Agmatine, a natural polyamine generated from arginine by arginine decarboxylase (ADC), has attracted increasing attention because of its pleiotropic beneficial effects on neuroprotection, lifestyle-related diseases, and gut-brain axis-mediated pathways. Although mammals appear to possess only limited capacity to synthesize endogenous agmatine, accumulating evidence suggests that agmatine derived from diet and the gut microbiota contributes to systemic levels of this polyamine. Previous studies have revealed that Aspergillus oryzae, the filamentous fungus foundational to traditional Japanese cuisine and indispensable for starch saccharification in sake, miso, soy sauce, mirin, and other fermented foods, produces high levels of agmatine specifically under solid-state cultivation. Subsequent studies identified a novel pyruvoyl-dependent ADC (Ao-ADC1) responsible for this unique agmatine production. This mini-review summarizes current knowledge on solid-state cultivation-specific agmatine production by A. oryzae, with a particular focus on the discovery and biochemical characteristics of Ao-ADC1. These findings challenge the commonly accepted notion that ascomycetes lack ADC. Understanding the molecular rationale and physiological significance of this unique agmatine biosynthetic pathway provides a foundation for rational strategies to enhance agmatine production in A. oryzae and for the development of agmatine-enriched fermented foods and nutraceuticals. Furthermore, integrating this fungal pathway with emerging insights into microbe-host interactions may further illuminate how fermentation-derived agmatine contributes to human health through gut-brain axis mediated mechanisms.
|

|
Scooped by
mhryu@live.com
Today, 1:23 AM
|
Here, we introduce Detectrons, modular biosensors that couples programmable toehold switches with retron-mediated reverse transcription to transduce RNA inputs into unique DNA barcodes. The ability to convert dynamic RNA signals into durable DNA records within living cells unlocks powerful new modes of transcript-based sensing with applications including viral infection detection. Through the construction of a synthetic toehold-retron library and application of machine learning, we uncovered key design principles that improve signal strength and specificity. We applied Detectrons to the multiplexed live-cell detection of specific phage infections, enabling transcript-triggered barcode synthesis and quantitative host susceptibility profiling in pooled bacterial populations. Detectrons are the first RNA-to-DNA transduction system, directly linking transient RNA detection to stable, sequence-encoded DNA outputs. This platform provides a scalable and generalizable strategy for phage screening and for recording transcriptional events in complex bacterial communities.

|
Scooped by
mhryu@live.com
Today, 1:15 AM
|
Conventional plant genome editing relies on tissue culture-mediated somatic cell regeneration, a technically demanding process that limits its application across diverse species. Emerging strategies now circumvent this bottleneck by enabling direct genome editing of meristematic or germline cells. Key advances include (i) genome editing via de novo meristem induction or dormant meristem activation; (ii) germline editing facilitated by graft-mobile tRNA-like sequence systems and haploid induction technologies; and (iii) optimized viral delivery platforms that exploit mobile RNA elements and compact editors such as TnpB to achieve efficient, transgene-free, heritable modifications across a broad range of genotypes and species. The development of robust, tissue culture-free editing platforms promises to revolutionize crop improvement pipelines and accelerate trait development for sustainable agriculture.

|
Scooped by
mhryu@live.com
Today, 1:06 AM
|
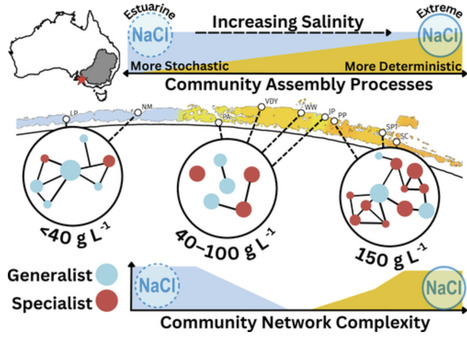
Coastal wetlands are highly vulnerable to climate-driven salinization, which reshapes critical microbial processes underpinning nutrient cycling and energy flow. We examined how sediment microbial communities vary with salinity across the Coorong Lagoon (South Australia), spanning estuarine (0–40 g L−1), intermediate (40–100 g L−1) and hypersaline (100–150 g L−1) waters. Salinity was found to be the dominant driver of sediment microbial community composition, diversity and assembly. High salinity favored specialists and homogenous community structures, with generalist bacteria persisting across intermediate salinities and supporting ecosystem resilience. Sulfur and carbon cycling is likely dependent on salinity, as bacterial sulfur-oxidizers were abundant estuarine specialists, whereas methane producers (Archaeal methanogens) and sulfate-reducers were enriched at high salinity. Deterministic microbial community assembly (homogeneous selection) was dominant, increasing at extreme salinity, which acted as a strong environmental filter. Community complexity increased at both high and low salinity ranges, with intermediate salinity exhibiting less complexity, suggesting community reorganisation under osmotic stress. The varied roles of specialists and generalists at different salinities support ecosystem function, where increased heterogeneity and specialization in hypersaline conditions suggest vulnerability of the community to disturbance. These findings provide insight into how microbially underpinned ecosystems may respond to future climate-driven salinization, important for making predictions and informing mitigation strategies.

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
The ability to engineer synthetic biomolecular condensates in living cells offers new opportunities to control intracellular organization, yet robust and programmable RNA-based systems have remained limited. Here, we introduce genetically encoded, modular platforms that generate RNA-driven condensates using nanostar-derived scaffolds. Systematic comparison of repeat-based and de novo designs identified nanostar variants that reliably assemble nuclear condensates in mammalian cells. Unexpectedly, condensate formation in cells is governed primarily by double-stranded RNA stems that recruit endogenous RNA-binding proteins, rather than by the kissing-loop interactions that drive assembly in vitro. This mechanistic shift highlights the divergence between cellular and in vitro environments and accounts for the limited orthogonality among scaffolds. Sequence refinement to reduce nonspecific pairing improved homotypic assembly and enhanced orthogonality. We further demonstrated functional compartmentalization by recruiting protein and RNA clients to modulate their stability and activity, and we incorporated an acyclovir-responsive allosteric switch to achieve reversible, small-molecule control of condensation. Together, this work establishes a versatile RNA-based toolkit for constructing programmable cellular compartments, advancing strategies for controlling RNA-protein organization and enabling new biosensing and therapeutic applications.

|
Scooped by
mhryu@live.com
Today, 12:40 AM
|
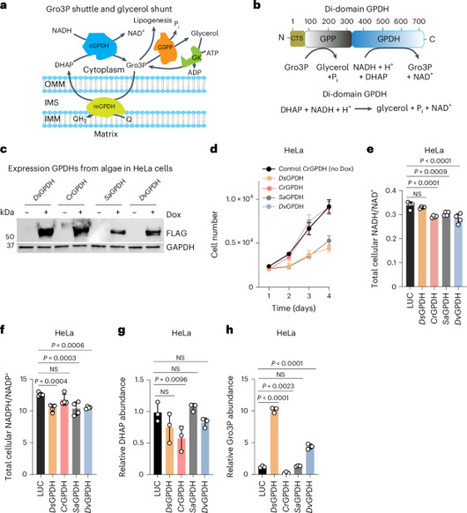
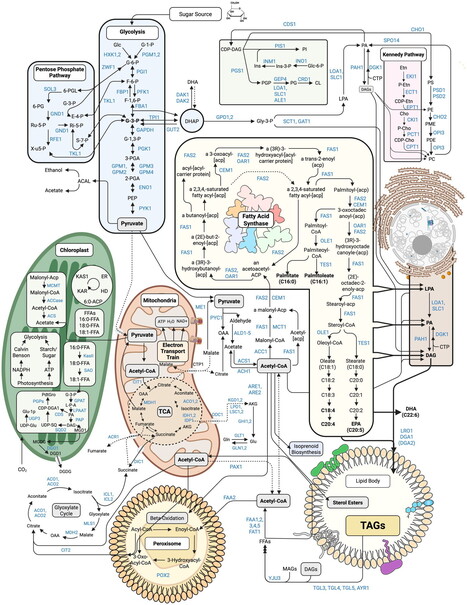
Dihydroxyacetone phosphate (DHAP), glycerol-3-phosphate (Gro3P) and reduced/oxidized nicotinamide adenine dinucleotide (NADH/NAD⁺) are key metabolites of the Gro3P shuttle, which transfers reducing equivalents between the cytosol and mitochondria. Targeted activation of Gro3P biosynthesis has recently emerged as a promising strategy to alleviate reductive stress. However, because Gro3P constitutes the backbone of triglycerides, its accumulation can promote extensive lipogenesis. Here we show that a genetically encoded tool based on a di-domain glycerol-3-phosphate dehydrogenase from the alga Chlamydomonas reinhardtii (CrGPDH) effectively operates both the alternative Gro3P shunt, which regenerates NAD⁺ while converting DHAP to Gro3P, and the glycerol shunt, which converts Gro3P to glycerol and inorganic phosphate, across transformed and primary mammalian cell cultures as well as mouse liver. CrGPDH expression supported proliferation of cancer cells under respiratory chain inhibition or hypoxia, as well as patient-derived fibroblasts with mitochondrial dysfunction. Moreover, CrGPDH decreased triglyceride levels in kidney cancer cell lines and reversed ethanol-induced triglyceride accumulation in mouse liver. Thus, CrGPDH represents a promising xenotopic tool to alleviate redox imbalance and associated impaired lipogenesis in conditions ranging from primary mitochondrial diseases to steatosis. The authors present a genetically encoded tool based on a bifunctional enzyme that can regenerate NAD+ while executing an engineered glycerol shunt. The tool successfully restored redox imbalance and modulated lipid metabolism in vitro and in a mouse hepatic steatosis model.

|
Scooped by
mhryu@live.com
Today, 12:31 AM
|
The demand for rapid and scalable biosensing technologies has motivated the development of antibody-free platforms capable of operating in complex sample environments. Here, we report an electrochemical biosensor based on engineered M13 bacteriophages displaying a SARS-CoV-2 spike S1–binding peptide immobilized on a reduced graphene oxide (rGO) transducer. The sensor employs a chemiresistive detection mechanism under a fixed low-voltage bias, enabling rapid electrical readout following target binding. Detection of S1 protein was achieved in buffer and in spiked complex matrices, including fetal bovine serum, pasteurized milk, and wastewater, demonstrating matrix tolerance under the tested conditions. The biosensor response is evaluated using a statistically defined binary detection criterion, with an operational limit of detection of 10⁻4 pg/mL in buffer. Compared to a previously reported antibody-functionalized rGO sensor fabricated using the same platform, the phage-based biosensor exhibits comparable sensitivity while offering advantages in genetic tunability and production scalability. While the present study focuses on proof-of-concept validation using spiked samples, these results highlight the potential of engineered phage–graphene interfaces as adaptable biorecognition elements for rapid electrochemical protein sensing in complex environments.

|
Scooped by
mhryu@live.com
February 16, 10:59 PM
|
Achieving neutral nitrogen (N) budgets is critical for soil health and sustainable agriculture. Thus, 31 studies were conducted for soybeans (Glycine max L.) throughout the United States, determining N inputs, biological N2 fixation via δ15N natural abundance method, and N outputs, seed N removal. On average, 53% of the N demand was met through N2 fixation, while the requirement of 62% indicates a significant shortfall for achieving neutral N budgets.

|
Scooped by
mhryu@live.com
February 16, 10:13 PM
|
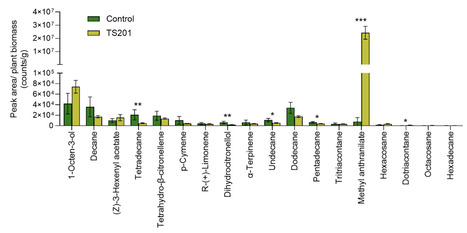
Microbial technologies are increasingly adopted to improve sustainable agriculture amid escalating economic, regulatory, and ecological pressures, yet few, if any, are supported by mechanistic understanding and are translated to products with demonstrated real-world performance and adoption. Pink-pigmented facultative methylotrophs (PPFMs) are known to promote plant growth and enhance pathogen resistance, but their impact on insect defense remains largely unexplored. Here we report on a PPFM species, Methylorubrum extorquens strain NLS0042, developed as TS201, a U.S. EPA–registered bioinsecticide that mitigates maize pest damage and improves yield. Across seven years (2016–2022) of field trials at 22 U.S. Midwest locations, TS201 increased maize yield by 220 kg/ha under moderate to high pressure of corn rootworms, the most destructive insect pest complex of maize in North America. Additionally, TS201 improved root architecture and reduced plant lodging. Large-scale on-farm evaluations across 81 sites in eight U.S. states (2023–2024) validated its agronomic benefit under diverse production conditions. Transcriptomic and volatile metabolite analyses revealed that TS201 induces anthranilate biosynthesis and accumulation of methyl anthranilate, a volatile compound that repels western corn rootworm larvae. Insect choice assays demonstrated significant larval avoidance of TS201-treated roots and confirmed the repellent effect of methyl anthranilate on corn rootworm larvae. These findings demonstrate that TS201 primes maize defense through multiple modes of action, offering a novel and effective microbial strategy to enhance crop resilience and support sustainable pest management.

|
Scooped by
mhryu@live.com
February 16, 5:21 PM
|
Biologically derived consumer oils and lipids are critical for the manufacture of an extensive array of products, including food, nutraceuticals, plastics, animal feed, cosmetics, pharmaceuticals, and fuels. However, a significant growth in demand has led to an unsustainable dependence on resource-intensive crops, further exacerbating deforestation, eutrophication, and the destruction of natural habitats. The ascendency of synthetic biology offers new opportunities to harness the potential of microbes as sustainable sources of microbial lipids. Recent advances in synthetic yeast genomics and the development of automated biofoundries could help secure the future of the sustainable lipid-based oil industry. This article outlines novel approaches to implement a sustainable future of consumer oils and lipids, which may help the transition to a practical microbial biomanufacturing economy.

|
Scooped by
mhryu@live.com
February 16, 5:15 PM
|
CRISPR-Cas systems confer prokaryotic immunity by integrating foreign DNA (prespacers) into host arrays. Type II-A systems employ Cas9 for protospacer-adjacent motif (PAM) recognition and coordinate with Csn2 and the Cas1-Cas2 integrase during spacer acquisition, yet their structural basis remains unresolved. Here, we report cryo-electron microscopy (cryo-EM) structures of the Enterococcus faecalis Cas9-Csn2-Cas1-Cas2 supercomplex in apo and DNA-bound states. The apo state (Cas9₂-Csn2₈-Cas1₈-Cas2₄) is a resting complex, while DNA binding forms a prespacer-catching complex threading DNA through Csn2’s channel, enabling Cas9 to interrogate the PAM sequence while sliding along the DNA. Cas9 and Csn2 jointly define a 30-bp DNA segment matching the prespacer length. Cas9 dissociation triggers structural reconfiguration of the Csn2-Cas1-Cas2 assembly. This exposes the PAM-proximal DNA, allowing Cas1-Cas2 to bind the exposed site for subsequent prespacer processing and directional integration. These findings reveal how Cas9, Csn2, and Cas1-Cas2 couple PAM recognition with prespacer selection, ensuring fidelity during adaptation.

|
Scooped by
mhryu@live.com
February 16, 3:52 PM
|
Flow cytometry (FC) is a widely used technique for analysing cells or particles based on the fluorescence of specific markers. Thresholds for fluorescence are typically set manually, a laborious, subjective process that scales poorly as FC technology advances. Machine learning (ML) methods can address these issues but often require technical expertise many bench scientists do not possess. Thus, accessible, open-source, and cross-domain ML-based FC tools are needed. We present AutoFlow, an easy-to-use, adaptable R Shiny application for automated flow cytometry (FC) analysis. AutoFlow supports two workflows: supervised and unsupervised learning. The application automates key preprocessing steps including fluorescence compensation, debris exclusion, single-cell identification, surface marker gating, MFI quantification, and downstream classification or clustering. Across three datasets, two publicly available (Mosmann and Nilsson Rare) and a novel bone marrow microphysiological system (BM-MPS) dataset, AutoFlow demonstrated robust performance. In the supervised workflow, multiclass classification on BM-MPS achieved 97.2% accuracy under a leave-one-timepoint-out scheme, with high sensitivity and specificity across major lineages. For rare populations, performance was strong: Mosmann Rare (0.03% prevalence) achieved 87.5% sensitivity, and 100% specificity, while Nilsson Rare (0.08% prevalence) achieved 87.9% sensitivity, and 99.9% specificity. The unsupervised workflow accurately grouped cells into biologically meaningful clusters, recovering known populations and identifying additional candidate populations with marker profiles consistent with true biology. AutoFlow offers a fast, reproducible, and scalable solution for FC analysis, enabling high-throughput studies and improving the discovery of rare or unexpected cell types.

|
Scooped by
mhryu@live.com
February 16, 1:01 PM
|
Temperature stress is a fundamental challenge for all organisms. While two-component systems (TCSs) are known to transduce environmental signals in microbes, their role in thermal sensing remains underexplored. Here, we unveil a novel thermosensitive TCS, DhqSR, in the thermophile Thermus thermophilus HB27. We demonstrate that the histidine kinase DhqS perceives thermal cues and autophosphorylates at His327. Its cognate response regulator, DhqR, is activated through a unique tyrosine-phosphorylation mechanism: phosphorylation at a unique Tyr84 residue, rather than the canonical aspartate. This atypical DhqSR system orchestrates cellular thermoadaptation by directly regulating a key enzyme in the shikimate pathway, type II 3-dehydroquinate dehydratase (DHQase). Our findings reveal a novel molecular mechanism of temperature sensing and adaptation, providing a new paradigm for microbial environmental adaptation and offering a unique toolbox for engineering thermotolerance.
|
 Your new post is loading...
Your new post is loading...






























provide simultaneous NAD+ recycling and efficient Gro3P clearance