Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:43 PM
|
Invasive pest Spodoptera frugiperda, known as the fall armyworm (FAW), evolves rapid resistance to chlorantraniliprole (CAP) via symbionts, reducing traditional control efficacy and causing economic losses. To address the formidable challenge of insecticide resistance, we introduce phage therapy into pest control, enabling precise targeting and efficient lysis of symbionts that mediate resistance. We employ zein to synchronously encapsulate phages and insecticides, constructing a nano-insecticide. This nano-insecticide ensures stability, exhibits robust performance by protecting phages against temperatures up to 60°C, and enhances their survival under UV irradiation by 83-fold. It intelligently responds to the pest gut enzymes for precise and controlled release, improving FAW control by 17% and overcoming resistance. Additionally, pesticide residue is reduced by 82.4%, with minimal impact on soil and maize microbial communities, preserving seedling growth. This modular, eco-friendly framework offers a sustainable solution for resistant pests, addressing the escalating challenge of resistant pests and paving the way for advancements in sustainable agriculture.

|
Scooped by
mhryu@live.com
Today, 12:09 PM
|
The biotransformation of acetaminophen, a pharmaceutical contaminant widely found in crop produce, in the rice phyllosphere highlights critical xenobiotic-plant-microbiota interactions. This study investigated acetaminophen uptake, translocation, and transformation in hydroponically exposed rice shoots, revealing accumulation at 8.33 ± 0.82 μg/g and conversion into hydroxylated, glycosylated, methylated, thiomethylated, sulfonated, dimerized, and amino acid-conjugated derivatives. Specifically, these transformations may be driven by plant enzymes (cytochrome P450, glycosyltransferases, sulfotransferases, and methyltransferases) and synergistically by enriched microbial genera (Sphingomonas, Pantoea, and Pseudomonas). Furthermore, acetaminophen stress altered the rice phyllosphere metabolome (elevated linoleic acid and jasmonic acid) and reshaped microbial communities, with enhanced degradation pathways and network complexity indicating adaptive stress mitigation. Overall, this integrated transcriptome, metabolome, and microbiome profiling provides mechanistic insights into the cooperative detoxification role of plant enzymes and phyllosphere microbes, offering perspectives on leveraging plant-microbiota interactions to reduce xenobiotic impacts on crops and food safety.

|
Scooped by
mhryu@live.com
Today, 11:51 AM
|
Adaptive, closed-loop control of cellular behavior is essential for next-generation therapies, yet most current treatments operate in an open-loop manner and lack robustness to patient variability and disease dynamics. Here, we establish a control-theoretic platform for rational engineering of closed-loop cell-based therapies that achieve precise and robust regulation. First, we introduce multi-dimensional nullgram profiling, a high-throughput approach that enables quantitative prediction and design of advanced genetic controllers in human cells across circuit topologies and parameter regimes in a single experiment. To evaluate dynamic therapeutic behavior, we next develop Cyberpatient-in-the-loop, an optogenetic digital twin platform that interfaces engineered mammalian cells with computational disease models, enabling systematic testing of closed-loop performance under realistic perturbations. Finally, we leverage these approaches to implement integral feedback cell therapies that sense inflammatory signals and autonomously regulate cytokine levels in primary immune cell cultures. Together, these results establish a general paradigm for engineering cell-based control systems and provide a foundation for next-generation cell therapies.

|
Scooped by
mhryu@live.com
Today, 10:32 AM
|
Large language models (LLMs) have shown remarkable success in natural language processing, prompting interest in their application to genomic sequence analysis. Genomic Language Models based on similar architectures offer a promising avenue for synthetic genome generation and characterization. However, their effectiveness for biological sequence modeling remains poorly characterized. We present a comprehensive evaluation of a state-of-the-art genomic Language Model (gLM), Evo 2, on multiple genomic reconstruction tasks. We tested Evo 2 on diverse prokaryotic, eukaryotic and viral genomes and assessed performance across key biological features and organizational patterns. Our results reveal systematic failures in gLM-based genomic reconstruction. While the synthetic sequences captured local sequence statistics, they consistently failed to preserve long-range genomic organization, repeat and k-mer composition, transcription factor binding site architecture, and evolutionary constraints. Generated sequences exhibited severe violations of natural genomic patterns and models showed particular difficulty with repetitive elements. These evaluation criteria provide a set of biologically grounded benchmarks for assessing the quality and realism of synthetic genomes. These findings suggest fundamental limitations in current gLM architectures for capturing the hierarchical, evolutionarily-constrained nature of genomic sequences. Our work highlights the need for specialized architectures that explicitly model biological constraints rather than relying solely on statistical patterns, with important implications for computational biology applications requiring realistic sequence generation.

|
Scooped by
mhryu@live.com
Today, 10:23 AM
|
Enhancing enzyme thermostability is crucial for industrial applications requiring robust performance under extreme conditions. Structure-based protein design models excel at improving thermal stability but often compromise enzymatic activity, while sequence-based models better preserve activity but struggle to enhance thermostability. It is challenging to efficiently generate multi-site mutants with both improved thermostability and intact activity using minimal experimental effort. Here, we used zearalenone hydrolase (RmZHD) and xylanase as model systems to evaluate different strategies for multi-site mutation design: (i) structure-based design with the ABACUS-R model, (ii) sequence-based design with the ProGen2 or MSA Transformer, (iii) integrated approaches combining either ProGen2 or MSA Transformer with ABACUS-R via Markov Chain Monte Carlo sampling with multi-objective scoring. Results showed that designing with ABACUS-R increased thermal stability by ~15°C but caused 80–100% activity loss. Sequence-based designs retained ~19% wild-type activity but failed to improve thermostability. Notably, zero-shot designs from integrating ABACUS-R with MSA Transformer achieved significant thermostability gains (ΔTm ~8°C) while preserving >95% wild-type activity. This highlights the potential of combining sequence-and structure-based deep learning models for developing industrially relevant thermostable enzymes.

|
Scooped by
mhryu@live.com
Today, 10:07 AM
|
Transcriptional regulation involves complex and dynamic protein–DNA interactions, which alter chromatin states and, consequently, regulate gene expression. In plants, current technologies face challenges in efficiently capturing dynamically DNA-binding proteins, especially transcription factors. Here, by leveraging the binding ability of dCas9 to specific DNA fragments and the labelling capacity of the TurboID protein for adjacent proteins, we have developed a CRISPR-based sequence proximity binding protein labelling system (CSPL) to detect promoter-binding proteins. Using this approach, we identified both known and novel upstream binding proteins on the PIF4 promoter in Arabidopsis, cabbage and rice. This demonstrates the powerful capabilities and broad potential applications of CSPL for detecting promoter-binding proteins in plants. The authors developed a CRISPR-based proximity labelling system to profile DNA-binding proteins in plants, especially the transcription factors of target genes, as exemplified by studies on the regulators of PIF4 in Arabidopsis, cabbage and rice.

|
Scooped by
mhryu@live.com
January 18, 12:37 PM
|
Drought stress severely constrains crop productivity, and while plant growth-promoting rhizobacteria (PGPR) are known to enhance drought tolerance by modulating host aquaporins (AQPs), the specific role of bacterial biofilm formation in this regulatory process remains poorly understood. Here, we demonstrate that biofilm formation is a pivotal mechanism through which Bacillus velezensis D103 confers drought resilience to maize. Under drought stress, maize root exudates synergistically enhanced D103 biofilm formation, which was essential for robust root colonization and mediated a drought-adaptive restructuring of the rhizosphere microbiome. Crucially, we found that an intact bacterial biofilm systemically upregulated key plant AQPs (ZmPIP2;6 and ZmTIP1;1), thereby enhancing root water transport capacity. Using virus-induced gene silencing, we further clarified the molecular mechanism underlying this biofilm-aquaporin link, revealing that ZmPIP2;6 is indispensable for D103-conferred drought tolerance. Our findings refine the current understanding of PGPR-mediated drought tolerance, highlighting that biofilms coordinate host AQP expression, rhizosphere microbiome assembly, and soil water retention to enhance drought resilience. This work provides a mechanistic basis for developing effective microbial inoculants.

|
Scooped by
mhryu@live.com
January 18, 12:11 PM
|
Spatial engineering has emerged as a transformative paradigm for orchestrating metabolic flux through biomolecular compartmentalization. In cellular systems, the cytosolic dispersion of heterologous enzymes and evolutionary-driven metabolic priorities of native pathways necessitate spatial solutions that transcend conventional enzyme engineering. Concurrently, in vitro metabolons provide critical mechanistic insights into enzymatic cascade reactions through controlled assembly. This review systematically evaluates several spatial engineering platforms for biocatalytic process control—including scaffolded compartments (liposomes, DNA origami, polymersomes, and bacterial microcompartments) and scaffoldless assemblies (membraneless organelles and coacervates)—designed to reconfigure metabolic landscapes in cellular or cell-free contexts. Through critical analysis of recent advances in model construction and functionalized applications, we establish a framework for understanding different spatial control principles governing pathway efficiency and flux redistribution. Finally, we conclude with a comprehensive assessment of current limitations in mechanistic elucidation, dynamic regulation and cross-system compatibility, while projecting future developments towards multifunctional spatial organization tools and biomimetic platforms for synthetic biology and cellular engineering.

|
Scooped by
mhryu@live.com
January 18, 11:40 AM
|
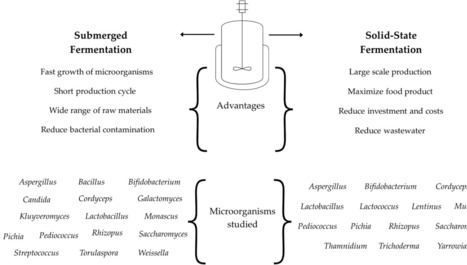
Filamentous fungi have emerged as ideal chassis cells for high-value products such as industrial enzymes, therapeutic proteins, and antibiotics, due to their broad substrate adaptability, efficient protein secretion capacity, and well-developed post-translational modification systems. However, the morphological characteristics of filamentous fungi during submerged fermentation present a significant challenge that cannot be overlooked in the biotechnology industry. This review systematically elaborates the fundamental role of polar growth and branching in hyphal morphogenesis and discusses the crucial impact of morphological regulation on fermentation performance. Through in-depth analysis of multi-level strategies, including process-based engineering control, genetic and cell wall modification approaches, and signaling pathway-mediated precise regulation, it clarifies the synergistic mechanisms underlying different regulatory methodologies. The rapid development of technologies such as high-throughput screening, genome editing, multi-omics sequencing, and artificial intelligence has enabled their integration into a collaborative engineering framework through functional complementarity and closed-loop data integration. This system, operating through a workflow of data-driven design, precise editing verification, and intelligent optimization iteration, will significantly enhance the efficiency and precision of morphological regulation. Such technological integration not only provides a systematic theoretical framework and technical guidance for understanding regulatory mechanisms and developing novel strategies, but also promotes the evolution of industrial fermentation toward intelligent and refined processes, thereby offering new technical pathways for green biomanufacturing.

|
Scooped by
mhryu@live.com
January 18, 11:31 AM
|
Internal ribosome entry sites (IRESs) provide compact RNA elements for noncanonical translation and hold promise as building blocks for RNA-based regulation in synthetic biology. However, the cricket paralysis virus (CrPV) IRES shows very low activity in Saccharomyces cerevisiae, limiting its broader utility despite extensive structural and biochemical studies. Here we report a yeast engineering strategy that enhances CrPV IRES-mediated translation by combining host modifications at three mechanistically distinct levels: translation initiation, tRNA modification, and mRNA stability. A reporter-based screen revealed host factors that influence IRES activity and uncovered a trade-off between IRES stimulation and maintenance of cap-dependent translation required for growth. Stepwise integration of nonsense-mediated decay deficiency, a tad3 temperature-sensitive allele, and wild-type eIF4E overexpression yielded a strain with up to an order-of-magnitude increase in reporter output compared with that of the parental strain. These results establish a proof-of-principle framework for host engineering of noncanonical translation.

|
Scooped by
mhryu@live.com
January 18, 10:36 AM
|
Microbial fermentation is a key biotechnological tool for producing bioactive metabolites such as alkaloids, carotenoids, essential oils, and phenolic compounds, among others, with applications in human health, agriculture, and food industries. This review comprehensively reviews recent information on the synthesis of valuable compounds and enzymes through fermentation processes. Here, we discuss the advantages of the different types of fermentation, such as submerged and solid-state fermentation, in optimizing metabolite production by bacteria, fungi, and yeast. The role of microbial metabolism, enzymatic activity, and fermentation conditions in enhancing the bioavailability and functionality of these compounds is discussed. Integrating fermentation with emerging biotechnologies, including metabolic engineering, further enhances yields and specificity. The potential of microbial-derived bioactive compounds in developing functional foods, pharmaceuticals, and eco-friendly agricultural solutions positions fermentation as a pivotal strategy for future biotechnological advancements. Therefore, microbial fermentation is a sustainable tool to obtain high-quality metabolites from different sources that can be used in agriculture, animal, and human health.

|
Scooped by
mhryu@live.com
January 18, 12:30 AM
|
Remote homology detection (RHD) is central to fold recognition and protein function annotation. While structural alignments provide a gold standard, they are computationally expensive. Encoding protein structures as sequences over structural alphabets offers a scalable alternative, but the relative performance of simple secondary-structure alphabets versus higher-resolution representations remains unclear. We systematically compare 20-letter (3Di), 8-letter (Q8), and 3-letter (Q3) structural alphabets across three large-scale fold recognition benchmarks of increasing difficulty, using both advanced and basic sequence alignment algorithms. All three alphabets perform close to structural alignment gold standards and substantially outperform sequence-based methods. Remarkably, the minimal Q3 alphabet, distinguishing only helices, strands, and loops, achieves robust performance. We further demonstrate the practical utility of this finding in a protein function annotation task for a newly sequenced genome. https://doi.org/10.6084/m9.figshare.c.8208161

|
Scooped by
mhryu@live.com
January 18, 12:22 AM
|
Soil pH is a predominant factor in structuring microbial communities; however, its role in shaping microbial life-history traits across large spatial scales remains underexplored. Here, we hypothesised that bacterial ubiquity, or niche breadth, across a diverse collection of soils is linked to genomic traits. We leveraged a national-scale survey of UK soils (the Countryside Survey) and 16S rRNA gene sequencing data with trait annotations (estimated genome size, coding density, and rRNA operon copy number) to examine trait-environment-niche breadth relationships. Our analyses revealed that soil pH was the dominant environmental driver of niche classification and bacterial community traits along the niche range. Low pH soils (pH <5.5) hosted ubiquitous taxa with larger genome sizes, lower coding densities and lower rRNA copy numbers, implying slower growing taxa with higher genetic facilities. Mildly acidic soils (pH 5.5 to 7) favor higher rRNA copy numbers, intermediate genome sizes and moderate coding densities. Alkaline soils (pH >7) feature communities with the smallest niche range, smallest genomes and highest coding densities. Here, specialization occurs through streamlining with simpler, smaller genomes favored. We found that generalist taxa were widespread across the pH range, becoming dominant under acidic conditions, while taxa adapted to higher pH were comparatively scarce in their distribution. These findings identify soil pH as a key physiological filter that aligns microbial genomic traits and ecological strategies across landscapes. By extending prior site-specific results to a broad-scale context, our study highlights how trait-based metrics can predict microbial responses to soil conditions, with implications for understanding ecosystem carbon cycling and informing land management practices aimed at sustaining soil health in the future.
|

|
Scooped by
mhryu@live.com
Today, 12:12 PM
|
Natural proteins often form intricate multidomain, oligomeric architectures. This presents a prima facie challenge to cellular homeostasis, as topologically complex proteins seldom refold efficiently in vitro. Here, we show that the efficient folding and assembly of the five-domain homotetramer β-galactosidase is obligatorily coupled to its synthesis on the ribosome, and we define the underlying mechanisms. During refolding from a denaturant, maturation of the catalytic domain is frustrated. Assembly outpaces monomer folding, and non-native oligomers accumulate. Efficient de novo folding is characterized by segmental domain folding, shaped by the binding of a nascent amphipathic helix to a cryptic pocket on uL23 on the ribosome surface. Homomer assembly also initiates cotranslationally via recruitment of a full-length subunit to the nascent polypeptide, and the failure to do so results in misassembly. Our results reveal how the ribosome can dictate the timing of folding and assembly to enable efficient biogenesis of a topologically complex protein.

|
Scooped by
mhryu@live.com
Today, 11:58 AM
|
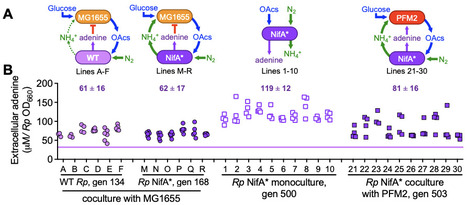
Some microbes externalize costly biosynthetic precursors in sufficient quantities to sustain a recipient population through cross-feeding. However, it is unclear whether metabolites are externalized purely for a reciprocal benefit or if metabolite externalization also plays a physiological role for the producer. Here we focus on adenine, a metabolite externalized by some strains of the phototrophic bacterium Rhodopseudomonas palustris at sufficient levels to support E. coli growth. In 10 long-term monocultures and 22 cocultures pairing R. palustris with E. coli, extracellular adenine externalized by all 140 isolates screened was 1.7 - 3.4-fold higher than that by the ancestor, suggesting that there was selective pressure for adenine externalization. We hypothesized that adenine is toxic to R. palustris. The CGA0092 growth rate decreased by half in the presence of about 0.3 mM external adenine. This inhibitory effect increased by an order of magnitude when we overexpressed adenine phosphoribosyltransferase to overcome a bottleneck in the purine salvage pathway, suggesting that toxicity stems from a metabolite derived from adenine. To assess whether adenine tolerance is connected to adenine externalization, we surveyed 12 evolved isolates and 49 environmental strains that externalized different levels of adenine, revealing a significant positive correlation. Our data suggests a physiological role for externalization of costly-metabolites like adenine at the origin of cross-feeding. In addition to cross-feeding, resulting metabolic interactions could be negative, considering that even a biosynthetic precursor like adenine can be inhibitory.

|
Scooped by
mhryu@live.com
Today, 11:45 AM
|
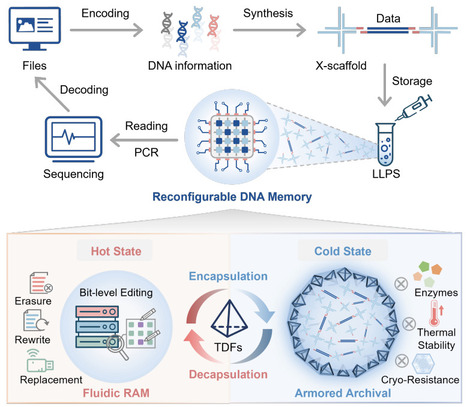
DNA has emerged as a promising medium for the post-silicon era of information storage due to its ultrahigh density and longevity. However, current systems are bifurcated, with solid-state systems providing robust cold archival but lacking accessibility, while fluidic molecular computing systems offer dynamic processing but suffer from low density and instability. This mutual exclusivity has hindered the development of hierarchical memory, a standard in modern computing, within molecular storage systems. Here, we bridge this gap by engineering a reconfigurable DNA memory architecture driven by programmable liquid-liquid phase separation (LLPS). Our system leverages sequence-based encoding to achieve an ultrahigh storage density of 7×10^10 GB/g, approaching the theoretical limits of DNA accessibility. In its fluidic hot state, DNA droplets enable rapid data loading (~83.8% in 5 min) and function as an in-memory editing platform supporting versatile, addressable bit-level operations including selective erasure (~65.1%) and high-efficiency rewriting and replacement (>99%) via programmable strand displacement. Importantly, to resolve the stability trade-off, we engineered a programmable phase transition whereby the triggered assembly of a rigid tetrahedral DNA framework (TDF) armor transforms liquid condensates into robust armored droplets. This cold state confers exceptional resistance to enzymatic and physical degradation, projecting multi-millennial data stability. By enabling reversible transitions between an editable, high-density computing mode and a stabilized archival mode, this work establishes the architectural foundation for scalable molecular information storage capable of hierarchical data management.

|
Scooped by
mhryu@live.com
Today, 10:25 AM
|
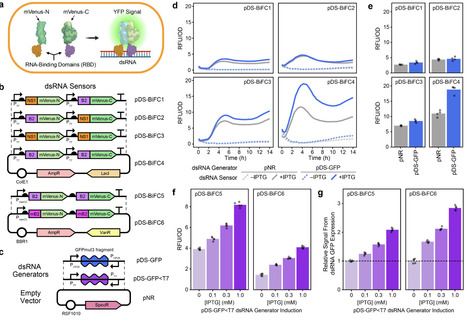
There is growing interest in engineering animal and plant microbiomes to deliver double-stranded RNA (dsRNA) for RNA interference (RNAi) applications. We developed a genetically encoded biosensor that uses bimolecular fluorescence complementation to monitor dsRNA levels within bacterial cells to accelerate the symbiont-mediated RNAi design-build-test cycle. We validated performance of the sensor in Escherichia coli and demonstrated enhanced dsRNA accumulation in engineered strains of the aphid symbiont Serratia symbiotica.

|
Scooped by
mhryu@live.com
Today, 10:15 AM
|
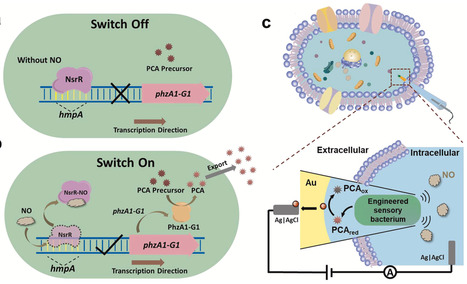
The intracellular gene circuit to transform biochemical signals to an electrical signal is the key for biosensor development to gap the information mismatch at the bio-electric interface, but a qualified gene circuit is difficult to design. In this study, we have shown the construction of an integrated synthetic gene circuit including the nitric oxide (NO)-responsive module and the phenylenediamine-1-carboxylic acid (PCA) synthesis module, to (1) equip a less renounced electroactive bacterium (e.g. Escherichia coli) with indirect electron transfer (IET) pathway to enhance electrical signal output, as well as (2) couple the IET gene circuit with the responsive gene circuit to sense small signal molecule, NO. The subsequent oxidation of PCA can be electrochemically quantified by the microelectrode, thereby establishing a signaling pathway from intracellular message to redox mediator, and finally to electrical signal output. In this way, the constructed microbial electrochemical biosensor for intracellular in situ NO analysis at the single-cell level owns high sensitivity, a wide linear detection range (100–2500 nM), and excellent selectivity. Therefore, this work shows an example for developing next-generation electroactive microorganisms (EAMs)-based electrochemical biosensors through synthetic biology tools with tailored and intelligent functionalities.

|
Scooped by
mhryu@live.com
January 18, 12:42 PM
|
Teosinte (Zea mays subsp. mexicana) has been proposed as a potential source of biological nitrification inhibition (BNI), yet how nitrogen (N) inputs modulate its exudate chemistry and associated nitrification processes remains unclear. We compared teosinte with three maize cultivars under N-deficient and N-replete conditions, integrating non-targeted metabolomics of root exudates, qPCR of rhizosphere amoA genes, and pure-culture assays with Nitrosomonas europaea. N fertilization enhanced total root exudation and reprogrammed the teosinte metabolome toward amino and phenolic acids, with histidine, glutamic acid, ferulic acid, and vanillic acid being markedly enriched. These compositional shifts coincided with reduced archaeal amoA abundance in teosinte (and Zhengdan958) but increased levels in Ye478 and Qi319. In culture, exudates from N-fed teosinte strongly inhibited N. europaea ammonia oxidation (~ 63%), whereas exudates from modern maize, except for Zhengdan958, showed little effect. Histidine, vanillic acid and ferulic acid reproduced inhibition in targeted assays, implicating them as candidate BNIs likely acting through copper chelation and phenolic interference. Collectively, these findings demonstrate that N availability reshapes teosinte exudate chemistry, thereby strengthening nitrification suppression through specific amino- and phenolic-acid release. Leveraging these wild traits could inform sustainable N management and enhance nitrogen-use efficiency in maize-based agroecosystems.

|
Scooped by
mhryu@live.com
January 18, 12:18 PM
|
Oral live microbial therapeutics (LMTs) show promise for halitosis, caries, and adjunctive periodontal care, yet benefits often fade after dosing stops. We synthesized evidence across indications and reframed development around quantifying and engineering persistence at intraoral sites, while outlining safety-by-design and delivery considerations for the oral niche. This narrative review integrated randomized trials, observational studies, and in vitro/ex vivo investigations to characterize clinical outcomes, persistence-related metrics, and engineering principles relevant to oral LMT development. Sources included PubMed/MEDLINE, Web of Science, Embase, and ClinicalTrials.gov, with backward/forward citation tracking. We included studies on LMTs in oral or gut contexts when mechanistically informative for oral applications (e.g., persistence, delivery, or biocontainment). Eligibility required clinical outcomes or persistence-related readouts. Two reviewers screened records and resolved disagreements by consensus. Reporting and assay principles were informed by STORMS and MIQE to support transparent, reproducible methods. Across indications, effects typically peak during dosing and attenuate after cessation, varying with strain, delivery format, and co-interventions (e.g., tongue dorsum debridement; standardized periodontal care). Persistence is rarely co-measured with clinical endpoints, limiting mechanistic interpretation. We outline a site-resolved measurement set, including time above the limit-of-detection, colonization area under the curve, apparent half-life (t½), and t½ under oral-mimetic shear, together with an engineering toolkit combining mucoadhesive/enamel-interactive carriers, single-cell coatings, and multilayer biocontainment (e.g., logic-gated/CRISPR kill switches, synthetic auxotrophy), and chemistry, manufacturing, and controls considerations. Embedding persistence metrics and safety-by-design into study protocols may support more durable outcomes, and standardized, site-resolved reporting will be essential for clinical translation.

|
Scooped by
mhryu@live.com
January 18, 12:00 PM
|
Genetically encoded biosensors provide powerful tools for coupling desired phenotypes to detectable outputs and have been extensively developed to detect a wide range of natural and unnatural products. When integrated with diverse high-throughput screening (HTS) approaches, these biosensors enable efficient product-driven screening across various throughputs, thereby expediting the engineering and optimization of microbial cell factories to produce various target compounds. For effective HTS of microbial cell factories, biosensors need to possess certain crucial characteristics. The performance features of biosensors significantly influence their application potential in HTS and can be precisely engineered through synthetic biology strategies. Furthermore, to ensure biosensor-driven HTS, additional engineering and optimizations of the biosensors are often required to increase the success rate and reduce false positives in the screening process. This review discusses the essential features of genetically encoded biosensors designed for HTS and then summarizes the latest advances in biosensor engineering for HTS purposes via synthetic biology strategies. Following this, the challenges and optimization of biosensors to adapt to different HTS processes are also discussed and exemplified. Finally, the key concerns and research prospects of developing biosensors for HTS applications are highlighted. Overall, this review provides comprehensive guidance on the engineering of genetically encoded biosensors and their applications in HTS for developing microbial cell factories to produce diverse target compounds.

|
Scooped by
mhryu@live.com
January 18, 11:33 AM
|
Actinomycetes represent a taxonomically and functionally diverse group of filamentous bacteria that are increasingly recognized for their role in environmental cleanup. Beyond their well-established capacity to produce antibiotics and other bioactive compounds, these microorganisms have shown remarkable potential in the degradation and transformation of a broad spectrum of pollutants, including petroleum hydrocarbons (e.g., crude oil, diesel), synthetic dyes (e.g., azo dyes), heavy metals (e.g., cadmium, lead), and pesticides (e.g., organochlorines). In recent years, significant progress has been made in uncovering novel actinomycete strains isolated from extreme and underexplored environments such as saline habitats, contaminated industrial sites, and marine sediments. These strains have demonstrated enhanced enzymatic activity and metabolic versatility in pollutant breakdown. Advances in molecular biology and omics-based techniques have further expanded our understanding of their biodegradative pathways and stress adaptation mechanisms. Nevertheless, several obstacles hinder their practical application, including inconsistent performance under field conditions, poor survival in competitive environments, and limited insight into their genetic regulation under contamination stress. This review not only summarizes current achievements (mainly from the last decade) but also highlights the urgent need for innovative strategies such as microbial consortia, metabolic engineering, and advanced formulation techniques to bridge the gap between laboratory findings and field-scale applications. Actinomycetes, therefore, stand as an untapped yet promising solution in the quest for efficient and eco-friendly bioremediation tools. bioremediation

|
Scooped by
mhryu@live.com
January 18, 11:26 AM
|
The release of labile organic carbon (OC) and nutrients during seasonal algal blooms can undermine blue carbon sequestration in coastal ecosystems. Although marine microorganisms mediate OC degradation during macroalgal decay, the underlying mechanisms remain poorly defined. This study employed an integrated multiomics approach (amplicon sequencing, metagenomics, and metatranscriptomics) to investigate microbial regulation of OC degradation and coupled nutrient cycling in coastal sediments with and without decomposing Sargassaceae. Total carbon in sediments increased by over 33% in the Sargassaceae area. Microbial α-diversity in the Sargassaceae area decreased significantly (p < 0.05), while processes linked to OC degradation, carbohydrate metabolism, nitrate (NO3–) reduction, inorganic phosphorus utilization, and sulfur metabolism were significantly upregulated (p < 0.05). Accordingly, gene expression and extracellular hydrolase activities targeting key biopolymers (i.e., cellulose, hemicellulose, starch, and chitin) were significantly upregulated (p < 0.05) in the area with Sargassaceae. Metabolism reconstruction of metagenome-assembled genomes identified Vibrio, Pseudoalteromonas, Alteromonas, and Exiguobacterium_A as primary OC degraders, with genomic capacities enriched in NO3– reduction and assimilatory sulfate reduction. Key environmental drivers─including the C/N ratio, dissolved organic carbon, total dissolved nitrogen (DON), and NO3–─shaped microbial metabolic activities during macroalgal decomposition. Our finding demonstrates that microbially driven OC degradation is a pivotal process coupled with nutrients cycling, advancing the mechanistic understanding of microbial carbon processing and its biogeochemical linkages during macroalgal decomposition in coastal ecosystems.

|
Scooped by
mhryu@live.com
January 18, 10:27 AM
|
Orphan genes - genes lacking detectable homologs outside a species - are widespread in microbial genomes and are thought to contribute to their adaptation and molecular innovation. However, not all predicted orphan genes may represent novel functional coding sequences. False positive orphan genes, also called spurious orphan genes, can arise from gene prediction errors. We reason that orphan genes lacking detectable expression are more likely to be spurious. To this end, we combined large-scale metatranscriptomic profiling of the human gut microbiome with machine learning to distinguish expressed orphan genes from spurious ones and to compare them with conserved genes found in multiple species. Using nearly 5,000 metatranscriptome libraries, we identified ~218,000 orphan genes supported by expression evidence, while ~330,000 predicted orphan genes lacked detectable expression, and were classified as spurious. We extracted 154 sequence, structural, and evolutionary features for each gene and trained XGBoost classifiers while accounting for genomic representation. The models achieved an area under the receiver operating characteristic curve (AUC) of 0.82 in distinguishing expressed orphan genes from spurious orphan genes and 0.93 in distinguishing expressed orphan genes from conserved genes. SHAP-based interpretation revealed clear biological signals. E.g., expressed orphans were present in more genomes than spurious ones and expressed orphan genes were shorter than conserved genes. This work improves orphan gene discovery and suggests that expressed orphan genes differ systematically from conserved genes and spurious orphan genes in sequence composition, structural constraints, and evolutionary signals.

|
Scooped by
mhryu@live.com
January 18, 12:24 AM
|
Life on Earth has long been regarded as homochiral, relying almost exclusively on a single enantiomer of sugars, typically the D-form. However, recent discoveries challenge this paradigm, including the identification of L-glucose-catabolizing bacteria and microbial L-glucoside hydrolases. Despite these findings, the metabolic diversity of organisms toward a broader range of atypical sugar enantiomers and their ecological relevance remains largely unexplored. This study aimed to identify and isolate microorganisms capable of catabolizing atypical enantiomers of diverse sugars. We performed enrichment cultures with either the D- or L-forms of glucose, fructose, xylose, and sorbose, using soil and activated sludge as microbial sources. Microbial growth was observed under all tested conditions, with the dominant taxa varying depending on the sugars supplied. Six phylogenetically distinct bacterial isolates exhibited the ability to catabolize atypical sugar enantiomers, two of which exhibited growth on all tested sugars. These findings uncover a previously unrecognized diversity in microbial sugar metabolisms, providing new insights into the environmental dynamics of atypical sugar enantiomers and offering a novel perspective on the principle of biological homochirality. Furthermore, this work lays a foundation for the development of biomanufacturing processes using racemic sugar mixtures synthesized via abiotic chemical reactions.
|
 Your new post is loading...
Your new post is loading...