 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:42 PM
|
Teosinte (Zea mays subsp. mexicana) has been proposed as a potential source of biological nitrification inhibition (BNI), yet how nitrogen (N) inputs modulate its exudate chemistry and associated nitrification processes remains unclear. We compared teosinte with three maize cultivars under N-deficient and N-replete conditions, integrating non-targeted metabolomics of root exudates, qPCR of rhizosphere amoA genes, and pure-culture assays with Nitrosomonas europaea. N fertilization enhanced total root exudation and reprogrammed the teosinte metabolome toward amino and phenolic acids, with histidine, glutamic acid, ferulic acid, and vanillic acid being markedly enriched. These compositional shifts coincided with reduced archaeal amoA abundance in teosinte (and Zhengdan958) but increased levels in Ye478 and Qi319. In culture, exudates from N-fed teosinte strongly inhibited N. europaea ammonia oxidation (~ 63%), whereas exudates from modern maize, except for Zhengdan958, showed little effect. Histidine, vanillic acid and ferulic acid reproduced inhibition in targeted assays, implicating them as candidate BNIs likely acting through copper chelation and phenolic interference. Collectively, these findings demonstrate that N availability reshapes teosinte exudate chemistry, thereby strengthening nitrification suppression through specific amino- and phenolic-acid release. Leveraging these wild traits could inform sustainable N management and enhance nitrogen-use efficiency in maize-based agroecosystems.

|
Scooped by
mhryu@live.com
Today, 12:18 PM
|
Oral live microbial therapeutics (LMTs) show promise for halitosis, caries, and adjunctive periodontal care, yet benefits often fade after dosing stops. We synthesized evidence across indications and reframed development around quantifying and engineering persistence at intraoral sites, while outlining safety-by-design and delivery considerations for the oral niche. This narrative review integrated randomized trials, observational studies, and in vitro/ex vivo investigations to characterize clinical outcomes, persistence-related metrics, and engineering principles relevant to oral LMT development. Sources included PubMed/MEDLINE, Web of Science, Embase, and ClinicalTrials.gov, with backward/forward citation tracking. We included studies on LMTs in oral or gut contexts when mechanistically informative for oral applications (e.g., persistence, delivery, or biocontainment). Eligibility required clinical outcomes or persistence-related readouts. Two reviewers screened records and resolved disagreements by consensus. Reporting and assay principles were informed by STORMS and MIQE to support transparent, reproducible methods. Across indications, effects typically peak during dosing and attenuate after cessation, varying with strain, delivery format, and co-interventions (e.g., tongue dorsum debridement; standardized periodontal care). Persistence is rarely co-measured with clinical endpoints, limiting mechanistic interpretation. We outline a site-resolved measurement set, including time above the limit-of-detection, colonization area under the curve, apparent half-life (t½), and t½ under oral-mimetic shear, together with an engineering toolkit combining mucoadhesive/enamel-interactive carriers, single-cell coatings, and multilayer biocontainment (e.g., logic-gated/CRISPR kill switches, synthetic auxotrophy), and chemistry, manufacturing, and controls considerations. Embedding persistence metrics and safety-by-design into study protocols may support more durable outcomes, and standardized, site-resolved reporting will be essential for clinical translation.

|
Scooped by
mhryu@live.com
Today, 12:00 PM
|
Genetically encoded biosensors provide powerful tools for coupling desired phenotypes to detectable outputs and have been extensively developed to detect a wide range of natural and unnatural products. When integrated with diverse high-throughput screening (HTS) approaches, these biosensors enable efficient product-driven screening across various throughputs, thereby expediting the engineering and optimization of microbial cell factories to produce various target compounds. For effective HTS of microbial cell factories, biosensors need to possess certain crucial characteristics. The performance features of biosensors significantly influence their application potential in HTS and can be precisely engineered through synthetic biology strategies. Furthermore, to ensure biosensor-driven HTS, additional engineering and optimizations of the biosensors are often required to increase the success rate and reduce false positives in the screening process. This review discusses the essential features of genetically encoded biosensors designed for HTS and then summarizes the latest advances in biosensor engineering for HTS purposes via synthetic biology strategies. Following this, the challenges and optimization of biosensors to adapt to different HTS processes are also discussed and exemplified. Finally, the key concerns and research prospects of developing biosensors for HTS applications are highlighted. Overall, this review provides comprehensive guidance on the engineering of genetically encoded biosensors and their applications in HTS for developing microbial cell factories to produce diverse target compounds.

|
Scooped by
mhryu@live.com
Today, 11:33 AM
|
Actinomycetes represent a taxonomically and functionally diverse group of filamentous bacteria that are increasingly recognized for their role in environmental cleanup. Beyond their well-established capacity to produce antibiotics and other bioactive compounds, these microorganisms have shown remarkable potential in the degradation and transformation of a broad spectrum of pollutants, including petroleum hydrocarbons (e.g., crude oil, diesel), synthetic dyes (e.g., azo dyes), heavy metals (e.g., cadmium, lead), and pesticides (e.g., organochlorines). In recent years, significant progress has been made in uncovering novel actinomycete strains isolated from extreme and underexplored environments such as saline habitats, contaminated industrial sites, and marine sediments. These strains have demonstrated enhanced enzymatic activity and metabolic versatility in pollutant breakdown. Advances in molecular biology and omics-based techniques have further expanded our understanding of their biodegradative pathways and stress adaptation mechanisms. Nevertheless, several obstacles hinder their practical application, including inconsistent performance under field conditions, poor survival in competitive environments, and limited insight into their genetic regulation under contamination stress. This review not only summarizes current achievements (mainly from the last decade) but also highlights the urgent need for innovative strategies such as microbial consortia, metabolic engineering, and advanced formulation techniques to bridge the gap between laboratory findings and field-scale applications. Actinomycetes, therefore, stand as an untapped yet promising solution in the quest for efficient and eco-friendly bioremediation tools. bioremediation

|
Scooped by
mhryu@live.com
Today, 11:26 AM
|
The release of labile organic carbon (OC) and nutrients during seasonal algal blooms can undermine blue carbon sequestration in coastal ecosystems. Although marine microorganisms mediate OC degradation during macroalgal decay, the underlying mechanisms remain poorly defined. This study employed an integrated multiomics approach (amplicon sequencing, metagenomics, and metatranscriptomics) to investigate microbial regulation of OC degradation and coupled nutrient cycling in coastal sediments with and without decomposing Sargassaceae. Total carbon in sediments increased by over 33% in the Sargassaceae area. Microbial α-diversity in the Sargassaceae area decreased significantly (p < 0.05), while processes linked to OC degradation, carbohydrate metabolism, nitrate (NO3–) reduction, inorganic phosphorus utilization, and sulfur metabolism were significantly upregulated (p < 0.05). Accordingly, gene expression and extracellular hydrolase activities targeting key biopolymers (i.e., cellulose, hemicellulose, starch, and chitin) were significantly upregulated (p < 0.05) in the area with Sargassaceae. Metabolism reconstruction of metagenome-assembled genomes identified Vibrio, Pseudoalteromonas, Alteromonas, and Exiguobacterium_A as primary OC degraders, with genomic capacities enriched in NO3– reduction and assimilatory sulfate reduction. Key environmental drivers─including the C/N ratio, dissolved organic carbon, total dissolved nitrogen (DON), and NO3–─shaped microbial metabolic activities during macroalgal decomposition. Our finding demonstrates that microbially driven OC degradation is a pivotal process coupled with nutrients cycling, advancing the mechanistic understanding of microbial carbon processing and its biogeochemical linkages during macroalgal decomposition in coastal ecosystems.

|
Scooped by
mhryu@live.com
Today, 10:27 AM
|
Orphan genes - genes lacking detectable homologs outside a species - are widespread in microbial genomes and are thought to contribute to their adaptation and molecular innovation. However, not all predicted orphan genes may represent novel functional coding sequences. False positive orphan genes, also called spurious orphan genes, can arise from gene prediction errors. We reason that orphan genes lacking detectable expression are more likely to be spurious. To this end, we combined large-scale metatranscriptomic profiling of the human gut microbiome with machine learning to distinguish expressed orphan genes from spurious ones and to compare them with conserved genes found in multiple species. Using nearly 5,000 metatranscriptome libraries, we identified ~218,000 orphan genes supported by expression evidence, while ~330,000 predicted orphan genes lacked detectable expression, and were classified as spurious. We extracted 154 sequence, structural, and evolutionary features for each gene and trained XGBoost classifiers while accounting for genomic representation. The models achieved an area under the receiver operating characteristic curve (AUC) of 0.82 in distinguishing expressed orphan genes from spurious orphan genes and 0.93 in distinguishing expressed orphan genes from conserved genes. SHAP-based interpretation revealed clear biological signals. E.g., expressed orphans were present in more genomes than spurious ones and expressed orphan genes were shorter than conserved genes. This work improves orphan gene discovery and suggests that expressed orphan genes differ systematically from conserved genes and spurious orphan genes in sequence composition, structural constraints, and evolutionary signals.

|
Scooped by
mhryu@live.com
Today, 12:24 AM
|
Life on Earth has long been regarded as homochiral, relying almost exclusively on a single enantiomer of sugars, typically the D-form. However, recent discoveries challenge this paradigm, including the identification of L-glucose-catabolizing bacteria and microbial L-glucoside hydrolases. Despite these findings, the metabolic diversity of organisms toward a broader range of atypical sugar enantiomers and their ecological relevance remains largely unexplored. This study aimed to identify and isolate microorganisms capable of catabolizing atypical enantiomers of diverse sugars. We performed enrichment cultures with either the D- or L-forms of glucose, fructose, xylose, and sorbose, using soil and activated sludge as microbial sources. Microbial growth was observed under all tested conditions, with the dominant taxa varying depending on the sugars supplied. Six phylogenetically distinct bacterial isolates exhibited the ability to catabolize atypical sugar enantiomers, two of which exhibited growth on all tested sugars. These findings uncover a previously unrecognized diversity in microbial sugar metabolisms, providing new insights into the environmental dynamics of atypical sugar enantiomers and offering a novel perspective on the principle of biological homochirality. Furthermore, this work lays a foundation for the development of biomanufacturing processes using racemic sugar mixtures synthesized via abiotic chemical reactions.

|
Scooped by
mhryu@live.com
Today, 12:18 AM
|
Global initiatives emphasize the need for harmonized soil biodiversity assessments. Efficient DNA extraction methods that accommodate larger soil volumes are essential for capturing higher trophic levels than bacteria and fungi and supporting extensive sampling campaigns. We developed and evaluated a scalable, cost-efficient, and automation-ready soil DNA isolation technique alongside commercial protocols. Three starting soil amounts (0.25 g, 2.5 g, and 5 g) were tested using widely used Qiagen kits, the developed isolation method, or combinations thereof. Lysis volumes ranged from 800 ul to 15 ml, and purification employed either silica membrane or carboxyl-coated magnetic beads. Four different types of soil, both agricultural and forest soil, samples were sequenced on an Illumina MiSeq platform using universal eukaryotic primers targeting the 18S rRNA SSU region, enabling detection of non-fungal eukaryotes such as soil mesofauna and protozoa. The developed protocol, which combined a tenfold increase in sample volume with hybrid purification steps, yielded the highest DNA recovery and consistently improved detected richness in several soil types. Species richness patterns varied by soil type and organism group: for eukaryotes and protozoa, commercial maxiprep methods along with the combination methods outperformed the miniprep approach in agricultural soils, while the developed technique excelled in coarse xeric forest soils. For metazoans, larger extraction volumes were associated with higher richness in forest soils. Our findings indicate that at least a tenfold increase in soil input compared to conventional 0.25 g is required to reliably capture mesofaunal diversity, with preliminary evidence suggesting further benefits at 20-fold volumes. We confirm that extraction volume is a key factor shaping detection of both soil metazoan and protozoan community compositions, with effects varying by soil type and organism group. The developed scalable approach offers a practical solution for large-scale soil biodiversity assessments, aligning with global monitoring goals and enabling integration of higher trophic levels into eDNA-based frameworks.

|
Scooped by
mhryu@live.com
January 17, 4:08 PM
|
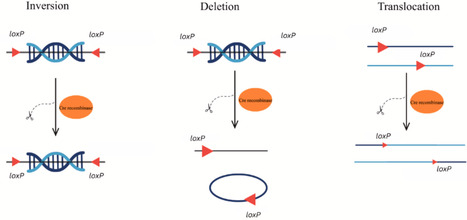
Saccharomyces cerevisiae, a model organism in genetics and molecular biology has been extensively engineered using various vector insertion techniques. This review compares and contrasts three prominent techniques: In vivo homologous recombination (HR), Cre-lox recombination and CRISPR/Cas9. In vivo HR leverages the organism's innate DNA repair machinery for easy vector integration and targeted genome modifications. Cre-lox recombination offers high specificity and efficiency at loxP sites, making it ideal for targeted gene excision or integration. CRISPR-Cas9 has revolutionized genome engineering with its precision and ability to target multiple loci simultaneously. Each technique has its strengths and limitations, including site dependency, off-target effects, and strain-specific variability. This review provides a comprehensive overview of these vector insertion techniques, highlighting their applications, advantages, and limitations in S. cerevisiae genome engineering and synthetic biology.

|
Scooped by
mhryu@live.com
January 17, 4:02 PM
|
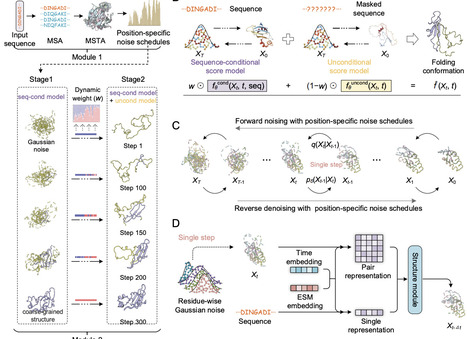
Despite remarkable advances in protein structure prediction, a fundamental question remains unresolved: how do proteins fold from unfolded conformations into their native states? Here, we introduce PathDiffusion, a novel generative framework that simulates protein folding pathways using evolution-guided diffusion models. PathDiffusion first extracts structure-aware evolutionary information from 52 million predicted structures the AlphaFold database. Then an evolution-guided diffusion model with a dual-score fusion strategy is trained to generate high-fidelity folding pathways. Unlike existing deep learning methods, which primarily sample equilibrium ensembles, PathDiffusion explicitly models the temporal evolution of folding. On a benchmark of 52 proteins with experimentally validated folding pathways, PathDiffusion accurately reconstructs the order of folding events. We further demonstrate its versatility across four challenging applications: (1) recapitulating Anton's molecular dynamics trajectory for 12 fast-folding proteins, (2) reproducing functionally important local folding-unfolding transitions in 20 proteins, (3) characterizing conformational ensembles of 50 intrinsically disordered proteins, and (4) resolving distinct folding mechanisms among 3 TIM-barrel proteins. We anticipate that PathDiffusion will be a valuable tool for probing protein folding mechanisms and dynamics at scale.

|
Scooped by
mhryu@live.com
January 17, 3:49 PM
|
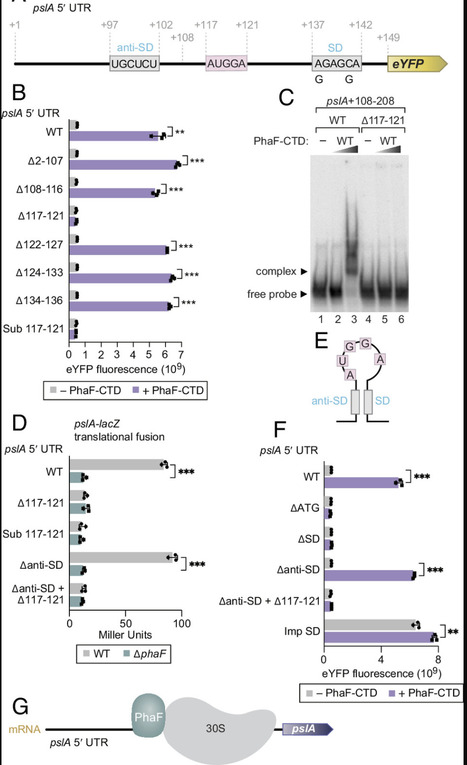
Bacterial RNA-binding proteins (RBPs) that control the translation of multiple transcripts act largely as negative regulators. Here, we report the identification and characterization of a positive regulator of translation (called PhaF) in the opportunistic pathogen Pseudomonas aeruginosa. Using CLIP-seq and CLAP-seq we identify upward of 50 transcripts targeted by PhaF. We demonstrate that PhaF acts to stimulate the translation of target mRNAs by binding upstream of the Shine–Dalgarno sequence using one or more of the multiple KPAA motifs located in an intrinsically disordered region of the protein. Importantly, we show that PhaF plays a key physiological role in P. aeruginosa through its translational control of the pslA transcript required for exopolysaccharide synthesis and biofilm formation. Our findings uncover an activator of translation in bacteria that binds target transcripts using an RNA-binding region reminiscent of those that are prominent in eukaryotic RBPs.

|
Scooped by
mhryu@live.com
January 17, 3:40 PM
|
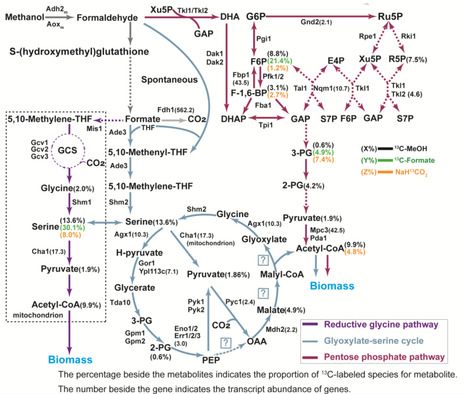
Methanol is a promising one-carbon (C1) feedstock for microbial bioconversion; however, engineered Saccharomyces cerevisiae often faces energetic constrains during its assimilation. Here, we develop SC-AOX25, an energy-efficient methylotrophic S. cerevisiae, through engineering of heterologous methanol-formaldehyde-formate (MFF) oxidation pathways coupled with adaptive laboratory evolution. SC-AOX25 efficiently generates adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide (NADH) during methanol metabolism while co-assimilating methanol-derived intermediates (formaldehyde, formate, and CO₂) via native glyoxylate-serine cycle, pentose phosphate pathway, and reductive glycine pathway. Key energy modules - Fdh1sc, Adh2m, Aoxm, and Rgi2m - are characterized for their roles in ATP/NADH synthesis and methylotrophic growth. Formaldehyde-induced DNA-protein crosslinks (DPCs) and large repeated DNA fragments suggest strategies for methanol detoxification and phenotype enhancement. Utilizing SC-AOX25, we enable CO₂ assimilation through non-native Calvin cycle during methanol fermentation, establishing the engineered strain as a robust and energy-efficient methylotrophic platform for further C1 engineering. Synthetic methylotrophic S. cerevisiae often faces energetic constrains during one-carbon assimilation. Here, the authors address this issue by engineering of heterologous methanol-formate-formaldehyde oxidation pathways to enable CO2 assimilation via non-native Calvin cycle during methanol fermentation.

|
Scooped by
mhryu@live.com
January 17, 3:29 PM
|
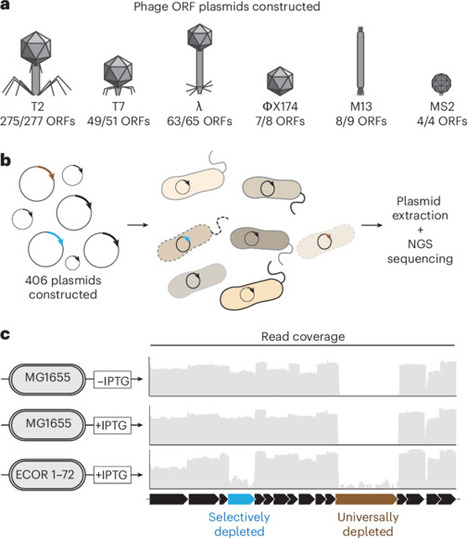
Bacteria have evolved sophisticated antiphage systems that halt phage replication upon detecting specific phage triggers. Identifying phage triggers is crucial to our understanding of immune signalling; however, they are challenging to predict. Here we used a plasmid library that expressed over 400 phage protein-coding genes from 6 phages to identify triggers of known and undiscovered antiphage systems. We transformed our library into 39 diverse strains of E. coli. Each strain natively harbors a different suite of antiphage systems whose activation typically inhibits growth. By tracking plasmids that were selectively depleted, we identified over 100 candidate phage trigger–E. coli pairs. Two phage proteins were further investigated, revealing that T7 gp17 and additional tail fibre proteins activated the undescribed antiphage system PD-T2-1 and identifying that λ gpE major capsid protein activated the antiphage system Avs8. These experiments provide a unique dataset for the continued definition of the molecular mechanisms underlying the bacterial immune system. A library of 400 phage protein-coding genes is used to find a trove of antiphage systems, revealing systems that target tail fibre and major capsid proteins.
|

|
Scooped by
mhryu@live.com
Today, 12:37 PM
|
Drought stress severely constrains crop productivity, and while plant growth-promoting rhizobacteria (PGPR) are known to enhance drought tolerance by modulating host aquaporins (AQPs), the specific role of bacterial biofilm formation in this regulatory process remains poorly understood. Here, we demonstrate that biofilm formation is a pivotal mechanism through which Bacillus velezensis D103 confers drought resilience to maize. Under drought stress, maize root exudates synergistically enhanced D103 biofilm formation, which was essential for robust root colonization and mediated a drought-adaptive restructuring of the rhizosphere microbiome. Crucially, we found that an intact bacterial biofilm systemically upregulated key plant AQPs (ZmPIP2;6 and ZmTIP1;1), thereby enhancing root water transport capacity. Using virus-induced gene silencing, we further clarified the molecular mechanism underlying this biofilm-aquaporin link, revealing that ZmPIP2;6 is indispensable for D103-conferred drought tolerance. Our findings refine the current understanding of PGPR-mediated drought tolerance, highlighting that biofilms coordinate host AQP expression, rhizosphere microbiome assembly, and soil water retention to enhance drought resilience. This work provides a mechanistic basis for developing effective microbial inoculants.

|
Scooped by
mhryu@live.com
Today, 12:11 PM
|
Spatial engineering has emerged as a transformative paradigm for orchestrating metabolic flux through biomolecular compartmentalization. In cellular systems, the cytosolic dispersion of heterologous enzymes and evolutionary-driven metabolic priorities of native pathways necessitate spatial solutions that transcend conventional enzyme engineering. Concurrently, in vitro metabolons provide critical mechanistic insights into enzymatic cascade reactions through controlled assembly. This review systematically evaluates several spatial engineering platforms for biocatalytic process control—including scaffolded compartments (liposomes, DNA origami, polymersomes, and bacterial microcompartments) and scaffoldless assemblies (membraneless organelles and coacervates)—designed to reconfigure metabolic landscapes in cellular or cell-free contexts. Through critical analysis of recent advances in model construction and functionalized applications, we establish a framework for understanding different spatial control principles governing pathway efficiency and flux redistribution. Finally, we conclude with a comprehensive assessment of current limitations in mechanistic elucidation, dynamic regulation and cross-system compatibility, while projecting future developments towards multifunctional spatial organization tools and biomimetic platforms for synthetic biology and cellular engineering.

|
Scooped by
mhryu@live.com
Today, 11:40 AM
|
Filamentous fungi have emerged as ideal chassis cells for high-value products such as industrial enzymes, therapeutic proteins, and antibiotics, due to their broad substrate adaptability, efficient protein secretion capacity, and well-developed post-translational modification systems. However, the morphological characteristics of filamentous fungi during submerged fermentation present a significant challenge that cannot be overlooked in the biotechnology industry. This review systematically elaborates the fundamental role of polar growth and branching in hyphal morphogenesis and discusses the crucial impact of morphological regulation on fermentation performance. Through in-depth analysis of multi-level strategies, including process-based engineering control, genetic and cell wall modification approaches, and signaling pathway-mediated precise regulation, it clarifies the synergistic mechanisms underlying different regulatory methodologies. The rapid development of technologies such as high-throughput screening, genome editing, multi-omics sequencing, and artificial intelligence has enabled their integration into a collaborative engineering framework through functional complementarity and closed-loop data integration. This system, operating through a workflow of data-driven design, precise editing verification, and intelligent optimization iteration, will significantly enhance the efficiency and precision of morphological regulation. Such technological integration not only provides a systematic theoretical framework and technical guidance for understanding regulatory mechanisms and developing novel strategies, but also promotes the evolution of industrial fermentation toward intelligent and refined processes, thereby offering new technical pathways for green biomanufacturing.

|
Scooped by
mhryu@live.com
Today, 11:31 AM
|
Internal ribosome entry sites (IRESs) provide compact RNA elements for noncanonical translation and hold promise as building blocks for RNA-based regulation in synthetic biology. However, the cricket paralysis virus (CrPV) IRES shows very low activity in Saccharomyces cerevisiae, limiting its broader utility despite extensive structural and biochemical studies. Here we report a yeast engineering strategy that enhances CrPV IRES-mediated translation by combining host modifications at three mechanistically distinct levels: translation initiation, tRNA modification, and mRNA stability. A reporter-based screen revealed host factors that influence IRES activity and uncovered a trade-off between IRES stimulation and maintenance of cap-dependent translation required for growth. Stepwise integration of nonsense-mediated decay deficiency, a tad3 temperature-sensitive allele, and wild-type eIF4E overexpression yielded a strain with up to an order-of-magnitude increase in reporter output compared with that of the parental strain. These results establish a proof-of-principle framework for host engineering of noncanonical translation.

|
Scooped by
mhryu@live.com
Today, 10:36 AM
|
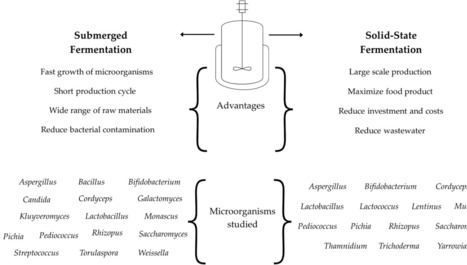
Microbial fermentation is a key biotechnological tool for producing bioactive metabolites such as alkaloids, carotenoids, essential oils, and phenolic compounds, among others, with applications in human health, agriculture, and food industries. This review comprehensively reviews recent information on the synthesis of valuable compounds and enzymes through fermentation processes. Here, we discuss the advantages of the different types of fermentation, such as submerged and solid-state fermentation, in optimizing metabolite production by bacteria, fungi, and yeast. The role of microbial metabolism, enzymatic activity, and fermentation conditions in enhancing the bioavailability and functionality of these compounds is discussed. Integrating fermentation with emerging biotechnologies, including metabolic engineering, further enhances yields and specificity. The potential of microbial-derived bioactive compounds in developing functional foods, pharmaceuticals, and eco-friendly agricultural solutions positions fermentation as a pivotal strategy for future biotechnological advancements. Therefore, microbial fermentation is a sustainable tool to obtain high-quality metabolites from different sources that can be used in agriculture, animal, and human health.

|
Scooped by
mhryu@live.com
Today, 12:30 AM
|
Remote homology detection (RHD) is central to fold recognition and protein function annotation. While structural alignments provide a gold standard, they are computationally expensive. Encoding protein structures as sequences over structural alphabets offers a scalable alternative, but the relative performance of simple secondary-structure alphabets versus higher-resolution representations remains unclear. We systematically compare 20-letter (3Di), 8-letter (Q8), and 3-letter (Q3) structural alphabets across three large-scale fold recognition benchmarks of increasing difficulty, using both advanced and basic sequence alignment algorithms. All three alphabets perform close to structural alignment gold standards and substantially outperform sequence-based methods. Remarkably, the minimal Q3 alphabet, distinguishing only helices, strands, and loops, achieves robust performance. We further demonstrate the practical utility of this finding in a protein function annotation task for a newly sequenced genome. https://doi.org/10.6084/m9.figshare.c.8208161

|
Scooped by
mhryu@live.com
Today, 12:22 AM
|
Soil pH is a predominant factor in structuring microbial communities; however, its role in shaping microbial life-history traits across large spatial scales remains underexplored. Here, we hypothesised that bacterial ubiquity, or niche breadth, across a diverse collection of soils is linked to genomic traits. We leveraged a national-scale survey of UK soils (the Countryside Survey) and 16S rRNA gene sequencing data with trait annotations (estimated genome size, coding density, and rRNA operon copy number) to examine trait-environment-niche breadth relationships. Our analyses revealed that soil pH was the dominant environmental driver of niche classification and bacterial community traits along the niche range. Low pH soils (pH <5.5) hosted ubiquitous taxa with larger genome sizes, lower coding densities and lower rRNA copy numbers, implying slower growing taxa with higher genetic facilities. Mildly acidic soils (pH 5.5 to 7) favor higher rRNA copy numbers, intermediate genome sizes and moderate coding densities. Alkaline soils (pH >7) feature communities with the smallest niche range, smallest genomes and highest coding densities. Here, specialization occurs through streamlining with simpler, smaller genomes favored. We found that generalist taxa were widespread across the pH range, becoming dominant under acidic conditions, while taxa adapted to higher pH were comparatively scarce in their distribution. These findings identify soil pH as a key physiological filter that aligns microbial genomic traits and ecological strategies across landscapes. By extending prior site-specific results to a broad-scale context, our study highlights how trait-based metrics can predict microbial responses to soil conditions, with implications for understanding ecosystem carbon cycling and informing land management practices aimed at sustaining soil health in the future.

|
Scooped by
mhryu@live.com
Today, 12:13 AM
|
Chronic exposure to inorganic arsenic remains a major global health concern, as arsenite is frequently present in contaminated food and drinking water and readily absorbed through the gastrointestinal (GI) tract. Once internalized, arsenite accumulates in tissues and contributes to long-term health effects, including cancer, organ dysfunction, and neurological disorders. Despite extensive efforts to reduce environmental contamination, there are currently no practical strategies to prevent dietary arsenite from entering the human body during digestion. Here, we report a synthetic biology-based approach that uses engineered probiotics to detect and sequester arsenite directly within the GI tract before systemic absorption occurs. We engineered E. coli Nissle 1917 (EcN), a probiotic strain, to function as a living arsenite-interception system. Central to this design is an arsenite-responsive genetic toggle switch that activates chelator expression upon exposure and sustains production under biostatic conditions, while automatically shutting off during active cell division to limit metabolic burden and enhance biosafety. In parallel, we engineered an arsenite-binding protein derived from the transcriptional regulator ArsR to eliminate DNA-binding activity while retaining high-affinity metal binding, yielding a non-toxic chelator suitable for intracellular sequestration. The resulting engineered strain efficiently removed arsenite from its surrounding environment in vitro while maintaining robust cell viability and growth. To translate these findings to an in vivo context, we developed a mass-transfer model describing arsenite distribution among the stomach lumen, engineered bacteria, and epithelial cells. This model guided the selection of a bacterial dose predicted to substantially deplete lumenal arsenite prior to epithelial uptake. Using this strategy, we demonstrated in a mouse GI model that oral administration of engineered EcN markedly reduced arsenite entry into the bloodstream compared with wild-type EcN or no-bacteria controls. Together, these results establish a programmable probiotic platform for intercepting dietary arsenite and highlight a potential strategy for preventing absorption of environmental toxicants using living microbial therapeutics.

|
Scooped by
mhryu@live.com
January 17, 4:05 PM
|
Enzyme engineering involves enhancing enzyme function and application through multidimensional technological systems. Its development encompasses elucidating sequence-structure-function relationships, exploring fitness landscapes, and multiscale regulation. Conventional enzyme engineering strategies include directed evolution (DE), rational/semi-rational design, residue co-evolution, and de novo design. DE mimics natural selection but is limited by high-throughput screening efficiency. Rational/semi-rational design integrates computational simulation with experimental validation to regulate enzyme performance. Residue co-evolution combines sequence co-evolution analysis and kinetic simulations, and de novo design is dedicated to achieving precise protein folding through physical modeling. In recent years, the breakthrough progress in machine learning (ML), especially deep learning (DL), has significantly enhanced the efficiency of all the above-mentioned methods; by accurately predicting mutational effects and efficiently exploring discontinuous sequence space, it provides powerful tools to improve or supplement the above strategies, thereby aiding in escaping local fitness optima traps. This review summarizes common strategies in enzyme engineering, including directed evolution, rational/semi-rational design, residue co-evolution, and de novo design, and further introduces the latest applications of ML and DL models in each of these fields. Although challenges persist, such as force field accuracy limitations, mutation sampling constraints, experimental throughput limitations, and epistatic effects, more comprehensive multimodal foundation models in the future are expected to integrate cross-scale parameters for intelligent design, and the establishment of standardized enzymology databases will enhance prediction reliability. Overall, AI-empowered enzyme engineering will drive a profound transformation toward a predictable and highly efficient pathway for enzyme design, providing more precise and powerful solutions for biocatalysis.

|
Scooped by
mhryu@live.com
January 17, 3:57 PM
|
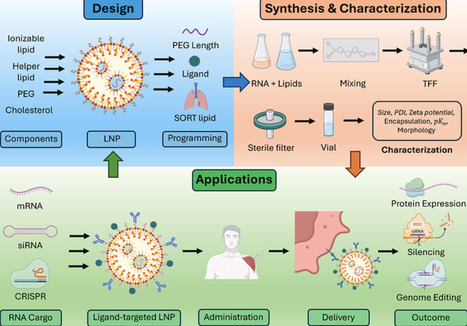
RNA therapeutics have come of age as clinically validated modalities including mRNA, siRNA, antisense oligonucleotides (ASOs), and in vivo genome editing, with lipid nanoparticles (LNPs) as the main non-viral delivery system. This review defines programmable LNPs as systems whose composition and interfacial chemistry are tuned to control organ tropism, cell specificity, intracellular trafficking, and immune interactions. We summarize design rules across four core components (ionizable lipid, phospholipid, cholesterol, PEG-lipid) and highlight levers like apparent pKa optimization (∼6–7 for hepatic delivery), biodegradable linkers, PEG-anchor-dependent shedding, ligands (e.g., GalNAc), and selective organ-targeting (SORT) lipids that redirect biodistribution beyond the liver. We survey advances in data-guided formulation, including DNA-barcoded in vivo libraries, machine learning, and physics-based prediction, plus scalable manufacturing (microfluidics, confined impinging-jet mixing, tangential-flow filtration) and Quality-by-Design with process-analytical technologies. A comprehensive characterization toolkit (size/ζ-potential, cryo-EM/SAXS, RNA encapsulation and integrity, apparent pKa, in vivo barcoding) maps to critical quality attributes. Applications span vaccines, protein replacement, siRNA/ASO delivery, and CRISPR platforms, with clinical examples like patisiran, COVID-19 and RSV mRNA vaccines, in-human transthyretin (TTR) editing, and individualized melanoma vaccination. We analyze translational constraints like endosomal escape, reactogenicity and anti-PEG immunity, complement activation, and lot-to-lot control, plus success factors: corona-aware design, dose-efficient potency at low lipid burden, redosing strategies, and fit-for-purpose biomarkers. Together, programmable LNPs offer a generalizable path to extrahepatic, cell-aware RNA medicine when coupled to rigorous analytics and platform manufacturing.

|
Scooped by
mhryu@live.com
January 17, 3:41 PM
|
Alternative splicing (AS) is a key mechanism for generating regulatory and phenotypic diversity in multicellular eukaryotes. Large-scale comparative transcriptomic studies have revealed that AS leads to lineage-specific and tissue-specific transcriptomic and proteomic changes, underscoring its contribution to the evolution of gene products and functions. In this review, we highlight the patterns and mechanisms of AS evolution across species, exploring how technological advancements are transforming our understanding of splicing evolution. Furthermore, we discuss mechanistic and functional insights from recent studies, including groundbreaking discoveries on how AS has shaped phenotypic evolution in mammals.

|
Scooped by
mhryu@live.com
January 17, 3:35 PM
|
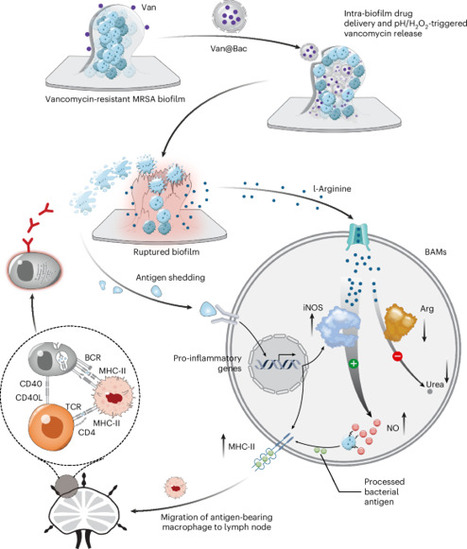
Bacterial biofilms, prevalent in human infections, present a major barrier to effective antibacterial therapy due to limited drug permeability and resistance. Here we introduce a ‘trick-bacteria-with-bacteria’ strategy that employs bacteria modified via calcium chloride treatment and antibiotic loading, followed by ultraviolet inactivation. These modified bacteria integrate selectively into biofilms of the same species, enabling targeted intra-biofilm drug release triggered by local pH and hydrogen peroxide. Species-specific integration is essential, as mismatched strains exhibit spatial segregation due to differences in surface adhesins and protein profiles. The strategy is effective against polymicrobial biofilms and demonstrated efficacy in treating biofilms formed by Staphylococcus aureus, Escherichia coli and Candida albicans. It also reinvigorates biofilm-associated macrophages by inducing the release of biofilm-derived l-arginine, enhancing immune responses. In vivo studies using subcutaneous and bone implant infection models showed stronger biofilm eradication and longer-term immunity in animals treated with modified bacteria compared with those treated with antibiotics, including resistance to re-infection. This approach could be adapted to modify infection-related bacteria from patients for personalized intra-biofilm drug delivery. Chemically modified and permeabilized bacteria allow for antibiotic delivery inside biofilms, inducing long-term immune protection and eradicating implant-related infections in preclinical models.
|






 Your new post is loading...
Your new post is loading...

















m-1str