Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:45 AM
|
Phage genome engineering methods accelerate the study of phage biology, the discovery of new functions, and the development of innovative genetic engineering tools. Here, we present QuickPhage, a rapid, technically accessible, precise, and cost-effective method for engineering Bacillus subtilis phages. Our approach uses CRISPR-Cas9 as a counter-selection system to isolate mutants of the model lytic siphovirus phage, SPP1. Efficient genome editing was achieved using homologous repair patches as short as 40 nucleotides, enabling streamlined patch construction and parallel engineering, resulting in highly accurate genome edits within a day. We applied QuickPhage to delete both essential and nonessential phage genes and to insert reporter genes. Protein production, such as GFP, was synthetically regulated using inducible systems without significantly affecting phage fitness, achieving induction levels of up to 400-fold. Time-series coinfection experiments with fluorescent protein expressing phages also revealed a highly efficient superinfection arrest mechanism that prevents reinfection as early as 13 min after initial infection. These findings highlight the potential of phages for protein production, opening new opportunities for metabolic engineering. This work also lays the foundation for systematic phage genome refactoring workflows and the development of phage-based tools for efficient DNA delivery, thereby expanding the synthetic biology toolbox for B. subtilis.

|
Scooped by
mhryu@live.com
Today, 12:09 AM
|
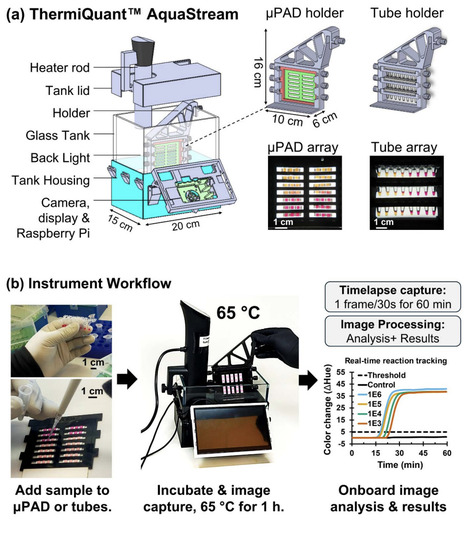
Microfluidic paper-based analytical devices (uPADs) are an attractive format for colorimetric nucleic acid amplification tests (NAATs) because they enable low-cost, portable diagnostics in resource-limited settings. However, researchers often optimize assays in liquid reactions in tubes before translating them to uPADs. Since both formats require separate instruments for incubation and real-time sensing, direct comparison of reactions between the two formats is difficult. To address these cross-platform limitations, we developed ThermiQuantTM AquaStream, a portable benchtop device (15 x 20 x 16 cm, ~5 kg; cost: USD 327) that supports seamless colorimetric loop-mediated isothermal amplification (LAMP) reactions in both uPADs and tubes under a common workflow. The system enables real-time reaction tracking (every 30 seconds) through onboard image processing, precise isothermal control (+/- 0.5 degree C) using a repurposed consumer-grade sous-vide heater, and medium-throughput (24 tubes or 42 uPADs). Testing with synthetic SARS-CoV-2 orf7ab DNA fragments demonstrated a limit of detection (LoD) of 50 copies per reaction in both formats (6.7 copies/uL for uPADs; 10 copies/uL for tubes). Standard calibration curves based on quantification time (Tq) and log10 DNA concentration showed strong full range linearity in tubes (from 1E6 to 50 copies/reaction, R2 = 0.98) and piecewise linearity in uPADs, with quantification reliability only above 1,000 copies/reaction (133 copies/uL, R2 = 0.99), which we set as the limit of quantification, LoQ. By unifying assay optimization across tube and uPAD formats, ThermiQuantTM AquaStream provides an affordable and versatile benchtop tool for molecular diagnostics for One Health applications in the clinic, on farms, and in the field.

|
Scooped by
mhryu@live.com
January 15, 11:36 PM
|
Type IV secretion systems (T4SS) are versatile molecular machines used by bacteria to secrete protein effectors into host cells, promoting pathogenesis, and to transfer DNA between bacteria through conjugation, driving horizontal gene transfer. Most, like Dot/Icm of the pathogen Legionella pneumophila or E. coli RK2, are primed for substrate delivery only upon contact with a target membrane, but mechanisms are unknown. A pilus could bind a receptor to initiate priming, but many T4SSs, especially those that deliver effectors, lack a pilus. Here, we present evidence that T4SSs are primed by direct contact with target membrane lipids. Combining fluorescence assays with genetics and biochemistry, we found that Dot/Icm drives lipid exchange between bacterial cells and between bacteria and synthetic membranes containing only lipids. Lipid exchange requires membrane contact but does not require ATP hydrolysis or even full complex assembly. Minimally, the outer membrane core complex protein DotG needs to be present in at least one of the apposed membranes. We similarly observed lipid mixing with the simpler E. coli RK2 T4SS, where we could follow lipid mixing and plasmid transfer simultaneously. We found that lipid mixing always preceded or accompanied plasmid transfer, suggesting it may be part of the contact-dependent priming mechanism. Lipid mixing was inhibited or promoted by lipids that inhibit or promote membrane fusion, respectively. Lipids inhibiting lipid mixing also inhibited substrate transfer. Together, our results suggest that initial contact between DotG outer segments and target membrane lipids promotes lipid mixing as part of the mechanism that primes T4SS for substrate translocation.

|
Scooped by
mhryu@live.com
January 15, 11:19 PM
|
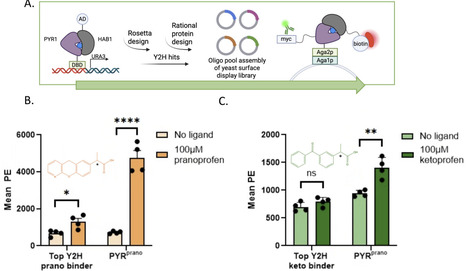
Non-steroidal anti-inflammatory drugs (NSAIDs) are pervasive environmental contaminants due to their frequent and widespread use, multiple paths of release into surface and ground water supply, diversity of the chemical class, and toxicity to aquatic and other non-target species. In particular, the 2-arylpropionic acid ("profen") class of NSAIDs poses significant risks to aquatic ecosystems due to incomplete removal during wastewater treatment. Current monitoring precludes high frequency testing at point sources. Here we present the engineering and application of a genetically encodable, protein-based biosensor for the detection of the NSAIDs ketoprofen and pranoprofen in wastewater effluent. We repurposed the plant hormone receptor PYR1 to bind selectively to profens using computational protein design, deep mutational scanning, and yeast 2 hybrid and yeast surface display screening. The resulting sensor, PYRNSAID, has a nanomolar limit of detection for ketoprofen and panoprofen, and micromolar sensitivity to the NSAIDs ibuprofen, fenoprofen, tolmetin and diclofenac. We also demonstrated dose responsive-activity of our sensor in simulated wastewater matrices containing the common wastewater contaminants sulfamethoxazole, caffeine, acetaminophen, and 2,4, dichlorophenol using a split Nanoluc luminescence assay. PYRNSAID is the first step towards a scalable, cost-effective alternative for real-time monitoring of pharmaceutical pollution.

|
Scooped by
mhryu@live.com
January 15, 8:46 PM
|
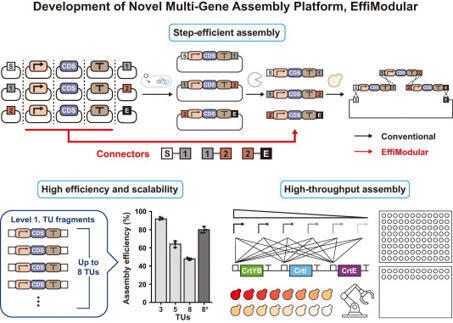
Rewiring the metabolic flux for efficient microbial conversion requires robust, scalable gene assembly. However, conventional gene assembly approaches are labor-intensive, highly experience-dependent, and require extensive expertise to ensure reproducibility and efficiency. Even with advanced automation platforms such as biofoundries, assembling gene arrays with multiple transcriptional units (TUs) remains challenging. In this study, we present Efficient Modular Gene Assembly (EffiModular), an integrated in vitro and in vivo gene assembly platform compatible with automated workflows. EffiModular enables the assembly of up to eight TUs with 80% efficiency in a single transformation. Integrated into a biofoundry workflow, it enabled the construction of 120 distinct yeast strains with varying levels of expression of the β-carotene biosynthesis genes within 3 days. Compared with conventional approaches, it significantly reduces procedural complexity, minimizes reliance on operator expertise, and accelerates workflow timelines. These features establish EffiModular as a next-generation gene assembly platform for scalable, reproducible gene assembly in biofoundry-based genetic engineering.

|
Scooped by
mhryu@live.com
January 15, 5:23 PM
|
U.S. select agent regulations ignore easily assembled DNA fragments, making synthesis screening ineffective regardless of accuracy. We acquired unregulated DNA collectively sufficient for a skilled individual to generate 1918 influenza from dozens of providers, demonstrating that fragments must be regulated as select agents.

|
Scooped by
mhryu@live.com
January 15, 5:07 PM
|
Direct reprogramming of immune cells holds promise for immunotherapy but is constrained by limited knowledge of transcription factor (TF) networks. Here, we developed REPROcode, a combinatorial single-cell screening platform to identify TF combinations for immune cell reprogramming. We first validated REPROcode by inducing type-1 conventional dendritic cells (cDC1s) with multiplexed sets of 9, 22, and 42 factors. With cDC1-enriched TFs, REPROcode enabled identification of optimal TF stoichiometry, fidelity enhancers, and regulators of cDC1 states. We then constructed an arrayed lentiviral library of 408 barcoded immune TFs to explore broader reprogramming capacity. Screening 48 TFs enriched in dendritic cell subsets yielded myeloid and lymphoid phenotypes and enabled the construction of a TF hierarchy map to guide immune reprogramming. Finally, we validated REPROcode’s discovery power by inducing natural killer (NK)-like cells. This study deepens our understanding of immune transcriptional control and provides a versatile toolbox for engineering immune cells to advance immunotherapy.

|
Scooped by
mhryu@live.com
January 15, 3:33 PM
|
Synthetic biology part discovery faces significant challenges due to inconsistent data organization and limited semantic search capabilities across existing repositories. We developed SynVectorDB, an embedding-based retrieval system that addresses these limitations through methodological innovations in data integration and AI-driven semantic search. Our approach integrates 19 850 biological parts from multiple sources (Addgene, iGEM Registry, laboratory collections), implementing systematic curation protocols that resulted in 7656 parts achieving verified status through literature-based validation and reliability assessment. We introduce a novel three-level hierarchical classification system organizing parts into functionally coherent categories (DNA Elements, RNA Elements, Coding Sequences, and Application Constructs) with detailed subcategorization. The core technical contribution employs BGE-M3 multilingual embeddings within a scalable vector database architecture to enable semantic similarity matching that significantly outperforms keyword-based retrieval methods. Standardized curation workflows enhance data comparability and search accuracy across heterogeneous sources. The dual deployment architecture ensures high performance through cloud services while maintaining open-source accessibility and deployment flexibility. The system maintains SBOL3 compatibility while providing innovative solutions for biological part organization and retrieval. Database URL: SynVectorDB is available in multiple deployment modes: web interface (https://svdb.sjtu.bio), local installation and source code (https://github.com/AilurusBio/synbio-parts-db), and MCP server integration for AI assistants (https://www.npmjs.com/package/synvectordb).

|
Scooped by
mhryu@live.com
January 15, 1:01 PM
|
Although natural sources of enzymes are limited to protein and RNA, some artificial DNAs exhibit catalytic activities. Representative functions of such DNAs, i.e. DNAzymes, are cleavage and ligation of nucleic acids. Here we developed a minimal DNAzyme with an RNA-cleaving activity by in vitro selection and secondary structure-based design. Its catalytic and substrate cores are only two and three nucleotides, respectively. This DNAzyme showed strict Zn2+ dependence at optimal pH 7.0–7.5. To elucidate its catalytic mechanism, we determined its three-dimensional structure by X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. The results consistently showed a B-DNA-like structure with base pairing and stacking throughout the molecule, unlike the kinked structures of larger DNAzymes. Notably, an A-base in the catalytic loop and a G-base in the substrate loop formed a non-Watson–Crick base pair. The catalytic Zn2+ coordinates to N7 of that G-base, enabling the Zn2+-hydrated water molecules to contacts O2′ and O5′ at the cleavage site. Considering that Zn(OH)+ and Zn2+ co-exist at the enzyme’s optimal pH, we propose a catalytic mechanism whereby these ions act as the base withdrawing H+ from O2′ and the acid donating H+ to O5′, generating the cleaved ends with 2′,3′-cyclic phosphate and OH groups.

|
Scooped by
mhryu@live.com
January 15, 12:57 PM
|
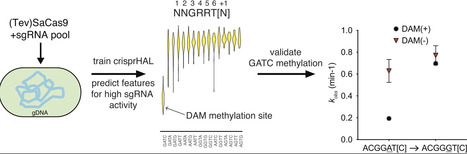
Cas9 nucleases defend bacteria against invading DNA and can be used with single guide RNAs (sgRNAs) as antimicrobials and genome-editing tools. However, bacterial applications are limited by incomplete knowledge of Cas9–target interactions. Here, we generated large-scale Staphylococcus aureus Cas9 (SaCas9)/sgRNA activity datasets in bacteria and trained a machine learning model (crispr macHine trAnsfer Learning) to predict SaCas9 activity. Incorporating downstream sequences flanking the canonical NNGRRN protospacer adjacent motif (PAM) at positions [+1] and [+2] improved predictive performance, with T-rich dinucleotides at these positions correlating with higher in vivo activity. Crucially, SaCas9 showed 10-fold reduced activity at sites containing a 5-NNGGAT[C]-3 PAM [+1] sequence in pooled sgRNA experiments in E. coli and Citrobacter rodentium. Plasmid cleavage assays in DNA adenine methyltransferase (DAM)-deficient E. coli confirmed that adenine methylation at GATC motifs inhibited SaCas9 activity. Removal of a DAM site within a PAM sequence enhanced cleavage, while introduction of a site reduced activity, directly linking adenine methylation to SaCas9 activity. These findings demonstrate that machine learning can uncover biologically relevant determinants of Cas9 activity. Avoidance of methylated PAMs may reflect an evolutionary adaptation by SaCas9 to discriminate self from nonself or to counter methylation as a phage and plasmid antirestriction strategy.

|
Scooped by
mhryu@live.com
January 15, 12:25 PM
|
Plants live in close association with microbial communities that support their health and growth. Previous research has indicated that the composition of these communities can differ between genotypes of the same plant species. Host-related factors causing this variation are mostly unknown. Microbiome genes, or M genes in short, are host genes that are involved in shaping the microbiome. We hypothesized that specific M genes are responsible for microbiome variation between rice genotypes and that it is connected to plant metabolites controlled by these genes. Our study was aimed at identifying plant metabolites driving genotype-specific microbiome assembly and establishing a link to host genetics. Targeted metabolite quantification was combined with microbiome profiling of the rice phyllosphere microbiome, association analyses on single-nucleotide polymorphism (SNP) level, and genetic modifications to validate microbiome-shaping effects of the discovered M genes. Targeted metabolite quantifications revealed that phenylpropanoid concentrations in rice leaves can substantially differ among 110 representative genotypes grown under the same, controlled conditions. Redundancy analyses (RDA) showed that these metabolites can explain 35.6% of the variance in their microbiomes. Further verification experiments resulted in the identification of two M genes. OsC4H2 and OsPAL06 are both plant genes with microbiome-shaping effects, mainly via their role in ferulic acid biosynthesis. Targeted gene mutation experiments confirm that distinct phyllosphere-associated bacterial groups are highly responsive to the discovered M genes. This study provides detailed insights into the links between host genetics and microbiome variation in plants. Knowledge about host genes that are in control of the microbiome paves the way for microbiome engineering and targeted plant breeding approaches.

|
Scooped by
mhryu@live.com
January 15, 12:58 AM
|
Genetically encoded pigments are powerful visual reporters and creative tools for biology, yet in plants the palette of pigment biosynthesis genes has remained largely limited to red betacyanins encoded by RUBY. Here we develop and characterize three new polycistronic constructs AMBER_v1, AMBER_v2, and GOLD that contain betalain biosynthesis enzymes to produce yellow, fluorescent betaxanthins in plant tissues. These tools expand the palette of publicly available pigmentation genes for use in plant research, education, and floral design.

|
Scooped by
mhryu@live.com
January 15, 12:49 AM
|
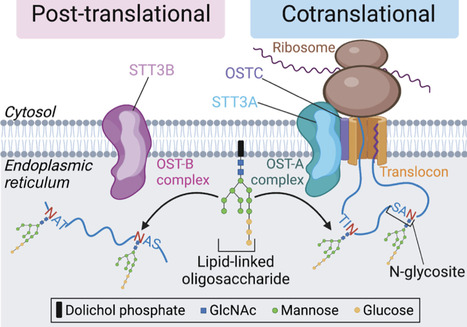
N-linked glycosylation in the endoplasmic reticulum (ER), catalyzed by two oligosaccharyltransferase (OST) complexes, has long been viewed as a constitutive post-translational modification. Recent discoveries suggest that OST complexes play a much more plastic and directive role in regulating ER processes. Here, we review this work and focus on one specific mechanism that uses N-glycosylation to regulate the stability of the ER chaperone HSP90B1. This degradative process regulates the cell-surface abundance of multiple signaling receptors that are HSP90B1 clients: toll-like receptors, WNT receptors, and growth factor receptors. This unusual system enables the status of ER-based processes to influence the sensitivity of cells to extracellular signals, with implications for tissue growth and development, inflammation, and immune function.
|

|
Scooped by
mhryu@live.com
Today, 12:41 AM
|
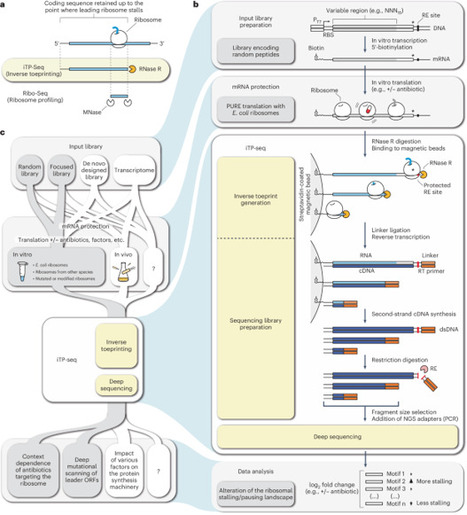
Uneven translation rates resulting from mRNA context, tRNA abundance, nascent amino acid sequence or various external factors play a key role in controlling the expression level and folding of the proteome. Inverse toeprinting coupled to next-generation sequencing (iTP-seq) is a scalable in vitro method for characterizing bacterial translation landscapes, complementary to ribosome profiling (Ribo-seq), a widely used method for determining transcriptome-wide protein synthesis rates in vivo. In iTP-seq, ribosome-protected mRNA fragments known as inverse toeprints are generated by using RNase R, a highly processive 3′ to 5′ RNA exonuclease. Deep sequencing of these fragments reveals the position of the leading ribosome on each mRNA with codon resolution, as well as the full upstream coding regions translated by these ribosomes. Consequently, the method requires no a priori knowledge of the translated sequences, enabling work with fully customizable transcript libraries rather than previously sequenced genomes. As a standardized framework for inverse toeprint generation, amplification and sequencing, iTP-seq can be used in combination with different types of libraries, in vitro translation conditions and data-analysis pipelines tailored to address a range of biological questions. Here, we present a robust protocol for iTP-seq and show how it can be integrated into a broader workflow to enable the study of context-dependent translation inhibitors, such as antibiotics. The time required to complete this workflow is ~10 d, and the workflow can be carried out by an experienced molecular biologist, with data analysis also requiring a working knowledge of command-line tools and Python scripts. This protocol describes inverse toeprinting coupled to next-generation sequencing, an in vitro approach to characterize bacterial translation at codon resolution that can accommodate custom synthetic libraries and various translation perturbations.

|
Scooped by
mhryu@live.com
January 15, 11:47 PM
|
Recirculating aquaculture system (RAS) effluents contain substantial nitrate and phosphate loads, posing a significant eutrophication risk to aquatic ecosystems if left untreated. While microalgal bioremediation is a promising strategy, expanding opportunities for valorisation is crucial for its successful implementation. This study couples the cultivation of nitrate-utilizing Chlamydomonas reinhardtii strain CC-1690 in RAS effluent with photobiological hydrogen (H2) production to achieve nutrient removal, bioenergy production, and high-quality biomass. Within 70 h, CC-1690 achieved near-complete depletion of nitrate and phosphate from the effluent. Subsequently, H2 production was induced via carbon limitation and anoxia, yielding a cumulative 49 umol H2 mg−1 Chl over 72 h. Although decreased chlorophyll content and Fv/Fm indicated physiological stress, key photosynthetic subunits (PsbA, PSBO, CP47, PetB and PsaA) remained largely stable. Importantly, the process preserved biomass quality; total lipid content increased slightly, enriched in palmitic (16:0) and α-linolenic (18:3ω-3) fatty acids while protein content was unaffected. These results demonstrate that integrating H2 production with RAS effluent bioremediation offers a robust circular economy approach, valorising wasted nutrients into bioenergy and high-quality biomass with potential downstream applications.

|
Scooped by
mhryu@live.com
January 15, 11:25 PM
|
Chemical conjugation to carrier proteins has been traditionally used to improve polysaccharides immunogenicity and to overcome the limitations of T-independent antigens, including lack of immunological memory and efficacy in infants. A double-hit approach, meaning that both polysaccharide and carrier protein belong to the same pathogen, may be particularly useful for targeting bacterial species with large glycan variability. Recently, bacterial protein glycosylation has been exploited to obtain glycosylated proteins in E. coli cytoplasm. In our work we have combined cytoplasmic glycoengineering and chemical conjugation for the development of novel selective glycoconjugates, with the aim to preserve the immunogenicity of the protein chosen as carrier. The potential protective protein MrkA, the major component of Klebsiella pneumoniae type 3 fimbriae, was successfully modified with a lactose moiety in E. coli. K. pneumoniae K2 K-antigen and O1v1 O-antigen were then covalently linked to MrkA at the level of this unique sugar handle and tested in vivo. Immune response against MrkA and sugars was evaluated in animal models. This work contributes to expand the application of the glycoengineering technology for the development of effective glycoconjugate vaccines.

|
Scooped by
mhryu@live.com
January 15, 11:16 PM
|
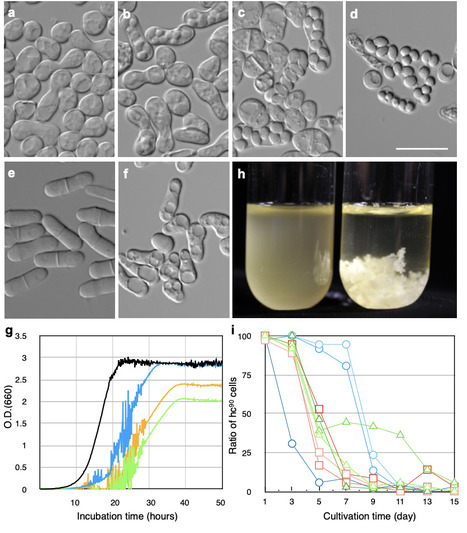
Sexual reproduction in yeast typically occurs under nutrient-limited conditions, producing stress-resistant spores that promote survival. However, wild Schizosaccharomyces japonicus strains isolated from fruit flies exhibit constitutive sporulation even under nutrient-rich conditions. Here we demonstrate that this nutrient-independent sporulation enhances survival by enabling spores to resist digestion by Drosophila melanogaster, and that although such strains are disadvantageous under nutrient-rich conditions, they are positively selected during insect predation. Genetic analyses reveal that multiple mutations in six meiotic regulatory genes, acting via epistasis, underlie this phenotype. This trait is widely distributed across natural populations throughout Japan. These findings suggest that constitutive sexual reproduction functions as an adaptive strategy facilitating insect-mediated dispersal, reflecting an ecological context where sexual reproduction is maintained despite its costs. This work provides new insights into yeast-insect symbiosis, evolutionary dynamics of sexual adaptation, and potential applications in yeast breeding.

|
Scooped by
mhryu@live.com
January 15, 8:31 PM
|
Understanding how cells make decisions over time requires the ability to link past molecular states to future phenotypic outcomes. We present TimeVault, a genetically encoded system that records and stores transcriptomes within living mammalian cells for future readout. TimeVault leverages engineered vault particles that capture mRNA through poly(A) binding protein. We demonstrate that the transcriptome stored by TimeVaults is stable in living cells for over 7 days. TimeVault enables high-fidelity transcriptome-wide recording with minimal cellular perturbation, capturing transient stress responses and revealing gene expression changes underlying drug-naive persister states in lung cancer cells that evade EGFR inhibition. By linking past and present cellular states, TimeVault provides a powerful tool for decoding how cells respond to stress, make fate decisions, and resist therapy.

|
Scooped by
mhryu@live.com
January 15, 5:22 PM
|
Emerging fungal pathogens have detrimental impacts on crops, animals, and humans. Despite the mounting threat of these emerging fungal pathogens, little is known about their transition from saprotrophic to pathogenic lifestyles. To gain insights into fungal lifestyle transitions, we study the Trichosporonales order, which includes both saprotrophic species and opportunistic human pathogens, as a system to reveal evolutionary adaptations leading to virulence in fungi. Here we use comparative genome analyses and experimental assays to demonstrate that the transition from saprotroph to opportunistic human pathogen is facilitated by adaptive translation. Codon optimization of metabolic genes grants these fungi the ability to quickly adapt to new environments. In this study, we link genomic data with fungal physiology, highlighting the role of adaptive translation in colonizing different environments and suggesting that gene translation optimization plays a critical role in fungal lifestyle evolution. Emerging fungal pathogens have detrimental impacts on crops, animals, and humans, however little is known about their transition to a pathogenic lifestyle. This study demonstrates that the transition from saprotroph to opportunistic human pathogen is likely facilitated by adaptive translation.

|
Scooped by
mhryu@live.com
January 15, 3:34 PM
|
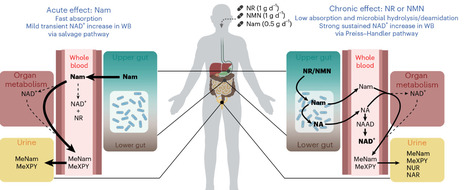
Nicotinamide adenine dinucleotide (NAD(H)) and its phosphorylated form NADP(H) are vitamin B3-derived redox cofactors essential for numerous metabolic reactions and protein modifications. Various health conditions are associated with disturbances in NAD+ homeostasis. To restore NAD levels, the main biosynthetic pathways have been targeted, with nicotinamide (Nam), nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN) being the most prominent boosters. However, while many preclinical studies have examined the effects of these precursors, a direct comparison in humans is lacking, and recent rodent research suggests that the NAD+-boosting effects of NR and NMN may depend on their microbial conversion to nicotinic acid (NA), a mechanism not yet confirmed in humans. Here we show in a randomized, open-label, placebo-controlled study in 65 healthy participants that 14 days of supplementation with NR and NMN, but not Nam, comparably increases circulatory NAD+ concentrations in healthy adults. Unlike the chronic effect, only Nam acutely and transiently affects the whole-blood NAD+ metabolome. Using ex vivo fermentation with human microbiota, we identify that NR and NMN give rise to NA and specifically enhance microbial growth and metabolism. We further demonstrate ex vivo in whole blood that NA is a potent NAD+ booster, while NMN, NR and Nam are not. Ultimately, we propose a gut-dependent model for the modes of action of the three NAD+ precursors with NR and NMN elevating circulatory NAD+ via the Preiss–Handler pathway, while rapidly absorbed Nam acutely affects NAD+ levels via the salvage pathway. Overall, these results indicate a dual effect of NR and NMN and their microbially produced metabolite NA: a sustained increase in systemic NAD+ levels and a potent modulator of gut health. ClinicalTrials.gov identifier: NCT05517122 . A comparison of the effects of different NAD+ boosters is lacking. This clinical study compares the efficacy of the NAD+ boosters NR, NMN and Nam in increasing circulating NAD+ levels and analyses their effects on gut microbial metabolism.

|
Scooped by
mhryu@live.com
January 15, 1:03 PM
|
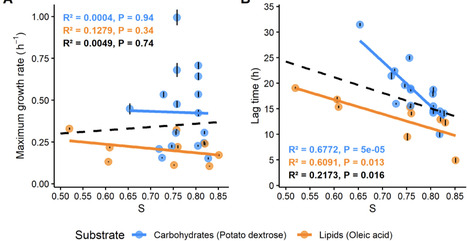
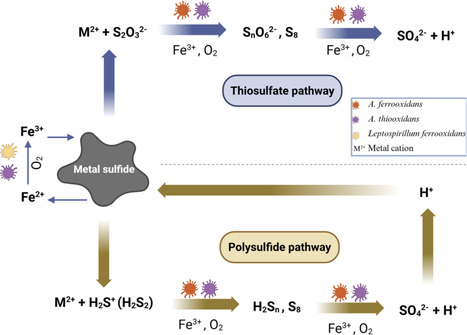
Heavy metals are essential for technological and economic growth but can cause serious environmental and health problems due to their toxicity and persistence. Traditional methods for metal recovery often have high costs and can create secondary pollution. Bioleaching offers a sustainable, low-energy, and eco-friendly alternative, effectively recovering metals from low-grade ores and various waste materials. Recovering metals from secondary sources such as industrial and electronic waste reduces the need for new mining, thus conserving natural resources and supporting circular economic goals. Recently, biomining has expanded beyond Earth, showing promising results in space environments. This review discusses the current understanding of bioleaching processes, their potential for sustainable metal recovery on Earth and in space, their challenges, and future perspectives. Overcoming technical challenges, such as raw material composition, slow reaction kinetics, optimization of process parameters, and addressing safety concerns is crucial. A further increase in research focus aiming at scaling up bioleaching technology is essential, alongside addressing ethical and economic concerns related to space mining.

|
Scooped by
mhryu@live.com
January 15, 12:58 PM
|
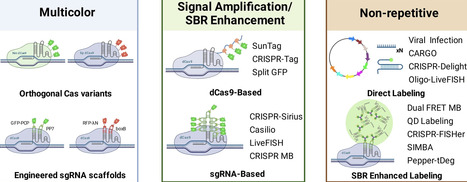
CRISPR-Cas-based live-cell imaging has rapidly become a central technology for studying genome dynamics with high specificity and flexibility. By coupling nuclease-deactivated Cas (dCas) with programmable guide RNAs, genomic loci can be tracked in living cells, providing direct insights into nuclear organization and chromatin behavior. While repetitive regions such as telomeres and centromeres are readily visualized, labeling non-repetitive loci remains more challenging due to weak signals and high background. Recent advances, including multicolor labeling strategies, innovative amplification systems based on dCas9 and single-guide RNA (sgRNA) engineering, and integration with novel fluorescent reporters, have markedly expanded the applicability of CRISPR imaging across the genome. These developments have expanded the multiplexing capacity of CRISPR imaging, improved signal-to-background ratios, and even enabled the visualization of non-repetitive genomic loci. Nonetheless, key challenges remain, including cellular toxicity, replication stress, and genomic instability associated with prolonged CRISPR expression. In this review, we summarize recent advances in CRISPR live-cell imaging and highlight key design trade-offs and biological constraints.

|
Scooped by
mhryu@live.com
January 15, 12:49 PM
|
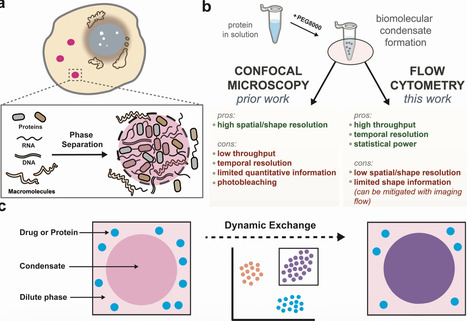
Biomolecular condensates are essential for cellular organization, yet their formation dynamics and molecular content exchange properties remain poorly understood. Here we show that flow cytometry provides a high-throughput, solution-based platform for analyzing condensate behavior at the single-droplet level. Using self-interacting NPM1 condensates as a model, we demonstrate that this approach quantifies phase behavior across protein and salt conditions, measures the partitioning of diverse macromolecules—including antibodies, lipids, small-molecule drugs, and RNA—and detects molecular colocalization with high statistical precision. Importantly, we establish a high-throughput assay to track real-time molecular exchange between preformed condensates and newly added, orthogonally tagged protein. These measurements reveal that condensate aging significantly reduces molecular dynamisms, likely due to altered biophysical properties with time. Compared to conventional imaging techniques that require surface immobilization or complex instrumentation, our method enables rapid, quantitative characterization of condensate dynamics and molecular content, providing a scalable framework for probing condensate function. He, Ongwae, and colleagues show that flow cytometry provides a high-throughput, solution-based platform for analysing the behavior of biomolecular condensates at the single-droplet level. Using this approach, the authors quantify condensate formation and track molecular exchange in real-time, and they reveal that condensate aging reduces molecular dynamisms.

|
Scooped by
mhryu@live.com
January 15, 1:12 AM
|
Small signaling peptides have emerged as central mediators of biological communication within and between species. In this review, we propose and define the concept of trans-kingdom peptides (TKPs) as short, bioactive peptides produced by one organism that exert specific physiological effects in another, often across taxonomic kingdoms. We summarize recent progress in identifying plant- and microbe-derived TKPs that function in symbiosis, parasitism, plant–microbe interactions, herbivory, and host-virus dynamics. TKPs modulate host defense, developmental programs, microbial community structure, and abiotic stress responses through highly specific interactions with conserved receptor systems. We highlight known peptide families mediating legume-rhizobia nodulation, nematode parasitism, and microbial immune suppression, as well as newly discovered viral- and insect-derived peptides that manipulate plant immunity. We discuss how they shape coevolutionary dynamics between hosts and interacting organisms. Finally, we outline current challenges and potential applications of TKPs in agriculture, biomedicine, synthetic biology, and environmental sustainability. Altogether, by framing their emerging properties and biological significance, we aim to provide a conceptual foundation and encourage interdisciplinary research into this expanding frontier of plant biology and inter-organismal communication.

|
Scooped by
mhryu@live.com
January 15, 12:51 AM
|
Plant natural products (PNPs) are specialized metabolites with diverse biological activities that are central to plant adaptation to biotic stresses. They act as chemical defenses against pathogens and pests, supporting plant survival under attack. Harnessing their potential as eco-friendly biopesticides requires a detailed understanding of their biosynthetic pathways and biological functions. This review highlights recent advances in the discovery and pathway elucidation of defensive PNPs and discusses strategies for their application through heterologous expression and enhanced in planta production.
|
 Your new post is loading...
Your new post is loading...