 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:10 AM
|
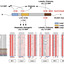
The specific partnership between legumes and rhizobia relies on a chemical dialogue. Plant flavonoids activate the bacterial transcription factor NodD, which triggers production of Nod factors that are recognized by the plant. Structural studies of the Pisum sativum (pea) symbiont Rhizobium leguminosarum NodD revealed two pockets that are essential for its activation by flavonoids. Comparative studies with NodD1 of Sinorhizobium medicae, the symbiont of Medicago truncatula, revealed that this specificity is determined by the shape of the pocket and by specific amino acids. A chimeric NodD containing the flavonoid recognition residues from S. medicae NodD1 in the R. leguminosarum NodD backbone was sufficient to complement nitrogen fixation in M. truncatula by an S. medicae nodD1 mutant, confirming the critical role of flavonoid recognition in host range.

|
Scooped by
mhryu@live.com
Today, 1:04 AM
|
Phosphorus (P) is a primary mineral nutrient essential for the growth and productivity of many crop plants. Although abundant in nature, its bioavailability is limited due to the prevalence of insoluble forms. An alternative for meeting agricultural P demand is the application of P-solubilizing microorganisms (PSMs), which mobilize it. Although progress has been made in the study of PSMs, knowledge gaps still exist regarding their role in sustainable agriculture. Therefore, this review examines the barriers to P acquisition in low-solubility soils and highlights recent advances in understanding the mechanisms of P solubilization mediated by plant-associated bacteria and fungi. The molecular strategies involved in the uptake and transport of P from soil in plants are also analyzed. Bacteria from genera such as Bacillus, Pseudomonas, and Streptomyces, as well as fungi including arbuscular mycorrhizal fungi, Aspergillus spp., and Penicillium spp., employ various approaches to solubilize P, leading to improved plant nutrition. These mechanisms, which include the production of organic acids, cation chelation, proton exudation, and phosphatase activity, can be inferred from experimental approaches or genome mining strategies. The role of PSMs as plant growth promoters and enhancers of plant nutrition across diverse environmental conditions are also discussed. Finally, we propose the integration of PSM consortia as multifunctional bioinoculants to promote sustainable agricultural practices.

|
Scooped by
mhryu@live.com
Today, 12:49 AM
|
The alpha-proteobacterium Zymomonas mobilis exhibits exceptional ethanologenic physiology, which makes it a traditional alcoholic beverage producer and a promising chassis for biofuel production. Although genetic tools for this organism have expanded in recent years, a fundamental aspect of its chromosome organization remains to be understood. In particular, Z. mobilis has been suggested to exhibit polyploidy, but this feature is not fully confirmed because of discrepancies among studies reporting the copy number of chromosomes. Here, we tagged the chromosome-partitioning protein ParB with a fluorescent marker to visualise its cellular localisation and estimate chromosome copy number in individual cells. Imaging showed that Z. mobilis exhibits several distinctive ParB foci throughout the cytoplasm and an accumulated focus at the pole, demonstrating that a single Z. mobilis cell contains at >5 copies of the chromosome at the oriC regions. After verifying its polyploidy, we sought to establish an efficient counter-selection system, which is crucial for engineering multiple copies of the chromosome. We assessed the efficacy of levan-sucrase (SacB) toxicity in Z. mobilis. We found that, despite Z. mobilis secreting a native extra-cellular sucrase SacB, heterologous periplasmically-localised Bacillus subtilis SacB rendered Z. mobilis cells sensitive to sucrose. We successfully used this effect for counter-selection when deleting and inserting targeted DNA sequences into the Z. mobilis genome. Together, this work provides important insights and tools for advancing Z. mobilis genetics and its biotechnological applications.

|
Scooped by
mhryu@live.com
Today, 12:42 AM
|
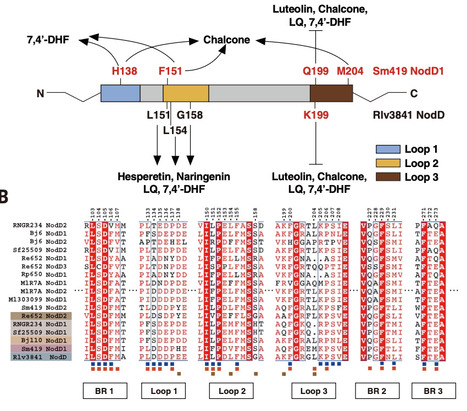
Automated free energy calculations for the prediction of binding free energies of congeneric series of ligands to a protein target are growing in popularity, but building reliable initial binding poses for the ligands is challenging. Here, we introduce the open-source FEgrow workflow for building user-defined congeneric series of ligands in protein binding pockets for input to free energy calculations. For a given ligand core and receptor structure, FEgrow enumerates and optimizes the bioactive conformations of the grown functional group(s), making use of hybrid machine learning/molecular mechanics potential energy functions where possible. Low energy structures are optionally scored using the gnina convolutional neural network scoring function, and output for more rigorous protein–ligand binding free energy predictions. We illustrate use of the workflow by building and scoring binding poses for ten congeneric series of ligands bound to targets from a standard, high quality dataset of protein–ligand complexes. Furthermore, we build a set of 13 inhibitors of the SARS-CoV-2 main protease from the literature, and use free energy calculations to retrospectively compute their relative binding free energies. FEgrow is freely available at https://github.com/cole-group/FEgrow , along with a tutorial. Automated free energy calculations for the prediction of binding free energies of ligands to a protein target are gaining importance for drug discovery, but building reliable initial binding poses for the ligands is challenging. Here, the authors introduce an open-source workflow for building user-defined congeneric series of ligands in protein binding pockets for input to free energy calculations.

|
Scooped by
mhryu@live.com
January 13, 6:31 PM
|
Optimization of flux distribution in central carbon metabolism is important to improve the microbial productivity. As the number of precursors required for synthesis differs for each target compound, optimal flux distribution also varies. A library of mutant strains with diverse flux distributions can aid in optimal strain screening. Therefore, in this study, we aimed to construct a library of Escherichia coli strains with stepwise changes in flux distribution by introducing mutations into the ribosome-binding sites of key enzyme genes on its chromosome. We focused on the flux ratios at the glucose-6-phosphate and acetyl-CoA branch points to enhance mevalonate production. Mutations were introduced into the RBS of pgi and gltA to vary the flux ratios of the two pathway branches. Furthermore, a combinatorial repression library comprising 16 strains was constructed by varying pgi and gltA expression at four levels, and a plasmid containing mevalonate synthesis genes was introduced into each strain. Batch cultures were performed to obtain strains with mevalonate titers and yields 2.4- and 3.4-fold higher than those of the parent strain. Overall, our combinatorial suppression library of pgi and gltA facilitated the effective identification of mutants with optimal metabolism for mevalonate production.

|
Scooped by
mhryu@live.com
January 13, 6:05 PM
|
Pre-mRNA splicing is a core process in eukaryotic gene expression, and splicing dysregulation has been linked to various diseases. However, very few small molecules have been discovered that can modulate spliced mRNA formation or inhibit the splicing machinery itself. This study presents a novel high-throughput screening (HTS) platform for identifying compounds that modulate splicing. Our platform comprises a two-tiered screening approach: A primary screen measuring growth inhibition in sensitized Saccharomyces cerevisiae (yeast) strains and a secondary screen that relies on production of a fluorescent protein as a readout for splicing inhibition. Using this approach, we identified 4 small molecules that cause accumulation of unspliced pre-mRNA in vivo in yeast. In addition, cancer cells expressing a myelodysplastic syndrome-associated splicing factor mutation (SRSF2P95H) are more sensitive to one of these compounds than those expressing the wild-type version of the protein. Transcriptome analyses showed that this compound causes widespread changes in gene expression in sensitive SRSF2P95H-expressing cells. Our results demonstrate the utility of using a yeast-based HTS to identify compounds capable of changing pre-mRNA splicing outcomes.

|
Scooped by
mhryu@live.com
January 13, 5:59 PM
|
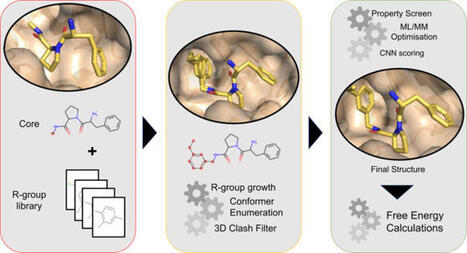
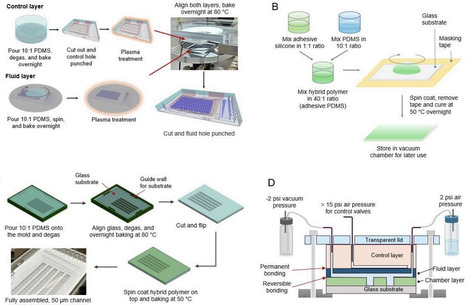
High-throughput microfluidics has transformed biomedical research by enabling precise and parallel sample handling, but most devices are single-use due to channel occlusion and contamination from experiments. Alongside low fabrication yield and reduced experimental success associated with dense microfeatures, this creates a major bottleneck for scalable high-throughput applications. We present a rapid, reusable, and modular high-throughput microfluidic platform with integrated microvalves for automation. The platform employs a multilayer architecture consisting of a custom casing, PDMS layers with dense microfeatures for fluid handling and culture, and a glass substrate. Permanent bonding is applied only between control and fluid layers, while reversible bonding is used at all other interfaces, including the substrate. Because substrate is the primary cell-contact surface and can be readily detached, the remaining layers can be disassembled, thoroughly cleaned, and reused with minimal processing on a new substrate. This approach improves repeatability and experimental success while reducing preparation time from days to ~2 hours. The disassemblable design also supports incorporation of application-specific layers between fluid layer and substrate, enhancing platform versatility for 3D culture. We validated performance through pressure/flow characterization and on-chip cell/organoid culture. Overall, our platform accelerates rapid high-throughput data generation across diverse biological applications.

|
Scooped by
mhryu@live.com
January 13, 4:00 PM
|
Harnessing heterosis has been important to agriculture, but its full potential remains constrained by the inability to clonally propagate hybrid seeds. Synthetic apomixis, engineering asexual seed formation in sexual crops, provides a path to fix heterosis through generations. This review compiles current advances in engineering synthetic apomixis, focusing on two major components: the substitution of mitosis for meiotic divisions and maternally derived embryo development by haploid inducer or parthenogenesis. Despite rapid progress made in rice, significant challenges remain for general implementation and extension to other crops. We discuss these challenges and the possible paths forward to make synthetic apomixis feasible for widespread application in agriculture.

|
Scooped by
mhryu@live.com
January 13, 3:47 PM
|
Understanding microbial growth and metabolism under controlled environments is critical for both fundamental research and bioprocess development. In this study, we present a cost-effective droplet-based microfluidic device enabling high-throughput screening of GFP expression in E. coli under varying glucose concentrations. Fluorinated Ethylene Propylene (FEP) tubing was selected for its low cost and compatibility with stable water-in-oil emulsification, facilitating robust droplet generation. The system achieved a 33333-fold reduction in media consumption compared to traditional Erlenmeyer flask cultures. Growth kinetics and GFP expression were assessed in both Erlenmeyer flasks and microdroplets, showing high qualitative correlation between platforms. Low glucose levels (5–10 g/L) supported rapid initial growth and early GFP production, followed by a fluorescence decline due to nutrient depletion. In contrast, higher glucose concentrations (25–50 g/L) prolonged the exponential phase and enhanced GFP production per unit biomass, though growth was slowed by overflow metabolism. In microdroplets, delayed GFP expression at 25 and 50 g/L were observed, and parallel bioreactor experiments confirmed that this delay is caused by oxygen limitation at high glucose concentrations. Importantly, the microfluidic device enables controlled variation of oxygen availability simply by adjusting droplet size or generation frequency, providing a powerful means to probe oxygen-sensitive metabolic behaviors. These results validate the microfluidic platform’s ability to mimic Erlenmeyer flask-scale dynamics, while uniquely allowing precise modulation of oxygen transfer conditions at the microscale. The system offers a reliable, miniaturized alternative for optimizing microbial bioprocesses with drastically reduced reagent use and increased experimental throughput.

|
Scooped by
mhryu@live.com
January 13, 3:21 PM
|
Food security and environmental sustainability require innovative agricultural approaches, with plant growth-promoting microbes offering promising solutions. This study explores the growth modulation of plants by Microbacterium sp. MB15 through secreted compounds, distinguishing between the effects of total diffusible substances and volatile organic compounds (VOCs). Microbacterium sp. MB15 exhibited a dose-dependent influence on wheat and Arabidopsis germination, growth, and root architecture, shifting from stimulation at low exposure levels to inhibition at higher concentrations. Total diffusible compounds, including auxin produced by the bacterium, exert their effect on Arabidopsis through the TIR1/AFB pathway and partially require the YUCCA genes. Microbacterium sp. MB15 VOCs, comprising ethanol, acetic acid, ethyl acetate, methanethiol and dimethyldisulfide, also elicited strong modulation of seed germination and seedling growth through mechanisms only partially dependent on TIR1/AFB and YUCCA. A proteomic analysis of seedlings exposed to bacterial VOCs (ProteomeXchange # PXD069087) indicated stimulation of carbohydrate and lipid mobilization, overexpression of proteins associated with a non-canonical auxin pathway, metabolism of indolic glucosinolates, defense responses, and sulfur-redox homeostasis. Additionally, high exposure to VOCs led to repression of genes for photosynthesis and chloroplast integrity. These findings help unravel the complex molecular responses underlying plant-microbe interactions.

|
Scooped by
mhryu@live.com
January 13, 2:26 PM
|
During the last few years, the body of data on proteins is expanding almost exponentially with the development of advanced methods for gene sequencing, protein structure determination, particularly by cryoelectron microscopy, and structure prediction using artificial intelligence-based approaches. These developments create the potential for a comprehensive exploration of the protein universe, the entirety of the proteins existing in the biosphere. Elucidation of the relationships among proteins including the most distant ones, where only the core fold is shared, is crucial for understanding protein functions, folding mechanisms, and evolution, as well as the evolution of cellular life forms and viruses. In this brief review, we discuss methods that shaped the field of protein bioinformatics, first, through comparative sequence analysis, and the recent developments in protein structure prediction that transformed the state of the art in the comparative analysis of distantly related proteins. The combination of the rapidly growing databases of genome and metagenome sequences with sensitive methods for sequence comparison and the new generation of structure analysis tools can make charting the protein universe at the structural level a realistic goal.

|
Scooped by
mhryu@live.com
January 13, 2:09 PM
|
Pseudomonas putida is a plant-beneficial rhizobacterium that encodes multiple type-VI secretion systems (T6SS) to outcompete phytopathogens in the rhizosphere. Among its antibacterial effectors, Tke5 (a member of the BTH_I2691 protein family) is a potent pore-forming toxin that disrupts ion homeostasis without causing considerable membrane damage. Tke5 harbors an N-terminal MIX domain, which is required for T6SS-dependent secretion in other systems. Many MIX domain-containing effectors require T6SS adaptor proteins (Tap) for secretion, but their molecular mechanisms of adaptor-effector binding remain elusive. Here, we report the 2.8 Å cryo-EM structure of the Tap3-Tke5 complex of P. putida strain KT2440, providing structural and functional insights into how effector Tke5 is recruited by its cognate adaptor protein Tap3. Functional dissection shows that the α-helical region of Tke5 is sufficient to kill intoxicated bacteria, while its β-rich region likely contributes to target membrane specificity. These findings delineate a mechanism of BTH_I2691 proteins for Tap recruitment and toxin activity, contributing to our understanding of a widespread yet understudied toxin family.

|
Scooped by
mhryu@live.com
January 13, 1:31 AM
|
Synthetic rewriting technologies, encompassing large-scale DNA assembly, transfer, maintenance, and rearrangement, enabled de novo synthesis or large-scale modifications of genomes. While significant progress has been made in model organisms of viruses, bacteria, and unicellular eukaryotes, their development in mammalian cells faces unique challenges. This review summarizes key breakthroughs in synthetic rewriting technologies, including megabase (Mb)-scale assembly of human DNA, yeast-mediated transfer methods, bottom-up human artificial chromosomes (HACs), and genome-scale rearrangement, along with emerging applications in constructing models and decoding genomes for mammals. These tools will expand functional engineering in mammals and deepen mechanistic insights into complex biological systems. Synthetic DNA rewriting technologies enable de novo synthesis or large-scale modification of genomes. Here the authors discuss key advances in the DNA rewriting toolbox at the level of large-scale DNA assembly, transfer, maintenance, and rearrangement, whilst highlighting emerging applications.
|

|
Scooped by
mhryu@live.com
Today, 1:07 AM
|
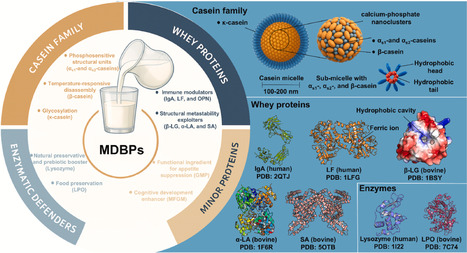
Milk-derived bioactive proteins (MDBPs) are essential for human growth, immune regulation, and neurodevelopment, possessing unique structure-function characteristics absent in plant-based and cultured proteins. Current industrial production adopts membrane separation, chromatography, and precipitation, but these methods face limited raw materials and high costs. Synthetic biology-enabled microbial cell factories (MCFs) offer a sustainable alternative, yet achieving both native-level bioactivity and high yield remains a substantial challenge. This review summarizes MDBPs from bovine and human milk, including their composition, abundance, structures, and physiological roles. It further delineates a complete recombinant production framework, encompassing chassis cell selection, transcriptional and translational optimization, post-translational modifications (PTMs), intracellular trafficking, secretion, and proteolytic stability. Particular attention is devoted to advanced microbial cell factory (MCF) strategies that integrate systems biology, multi-omics analytics, and protein engineering to address production bottlenecks, thereby providing a coherent technical roadmap for the industrial translation of MDBP research. MCFs provide a sustainable platform for MDBP manufacturing, offering high process controllability and scalability. Notably, achieving precise recapitulation of complex PTMs and optimizing secretion pathways are key to ensuring higher bioactivity and yield. Future artificial intelligence (AI)-assisted design-build-test-learn (DBTL) cycles will further improve MCF engineering and broaden MDBP applications in functional foods, clinical nutrition, and targeted therapeutics.

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
Bacterial chromosomes contain Genomic Islands carrying genes involved in virulence, mutualism, and resistance to antibiotics that are transferred horizontally. Some of them conjugate autonomously (Integrative Conjugative Elements, ICEs), while others use relaxases to hitch on the former's conjugation machinery (Integrative Mobilizable Elements, IMEs). Yet, the mobility mechanism of most Genomic Islands remains elusive. Here, we explore the hypothesis that many carry origins of transfer (oriTs) by conjugation. Since very few known oriTs were found in ICEs and IMEs, we identified 52 novel families of oriTs that cover most integrative elements in six major nosocomial species. Most of these oriTs are specific to integrative elements, suggesting a clear split in the targets of hitchers of conjugative elements between plasmids and integrative elements. Among 7,363 Genomic Islands, we discovered more than 1,500 IMEs carrying an oriT and lacking relaxases and MPF systems. These elements, coined iOriTs, form diverse large ancient families that can be found in different species. These hitchers are genetically unrelated to the putative helpers, beyond the similarity at the oriT sequence, and often outnumber them. They include well-known pathogenicity islands, for which the mechanisms of mobility remained unknown. Unlike plasmids, iOriTs have few antibiotic resistance genes, but high density of virulence factors. Like plasmids, the vicinity of oriTs concentrates defense and counter-defense systems potentially favoring Genomic Islands dissemination. Hence, iOriTs are frequent integrative mobile genetic elements that evolved to transfer horizontally by hitching on other elements while contributing to bacterial genetic adaptation.

|
Scooped by
mhryu@live.com
Today, 12:47 AM
|
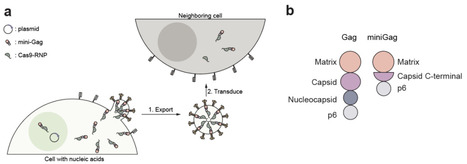
Genome editing enzymes have vast therapeutic potential. However, achieving sufficient delivery in vivo remains a major challenge, because editing machinery is confined to the subset of transfectable cells in a tissue. Here, we tested the possibility that genome editing could be amplified in vivo by programming transfected cells to produce and transfer editing enzymes in lipid vesicles to neighboring cells. Our data show that this NANoparticle-Induced Transfer of Enzyme (NANITE) strategy tripled editing efficiency in cultured cells relative to non-spreading controls. Furthermore, a single intravenous injection of the NANITE plasmid into mice induced ~3-fold higher levels of liver editing at the Ttr locus relative to non-spreading controls, with corresponding reductions in serum transthyretin levels. Amplifying therapeutic enzymes in situ offers a nonviral and non-infectious strategy to overcome low delivery efficiencies and reduce effective dose requirements.

|
Scooped by
mhryu@live.com
January 13, 11:59 PM
|
Bacterial cellulose (BC) is produced by diverse bacterial species, including members of the genus Komagataeibacter, and has emerged as a potential sustainable alternative to conventional materials such as plastics and leather. Its production by bacteria is rapid and easily scalable, and via genetic modifications or culturing the bacteria with other microbes, it is possible to rationally enhance the BC that is produced, for example by adding functional components. Here, we systematically evaluated BC functionalization strategies, including genetic engineering and co-culturing with other engineered microbes across the most widely used Komagataeibacter species: K. rhaeticus, K. xylinus, K. medellinensis and K. sucrofermentans. We established that all tested strains are amenable to DNA transformation and capable of expressing heterologous genes from identical genetic constructs. However, we identified species-specific differences in heterologous gene expression, cellulose production in varying environmental conditions and in the abilities of the strains to co-culture with Escherichia coli. Additionally, we demonstrated that all these cellulose-producing bacteria can establish functional symbiotic co-cultures with yeast. While inter-species variations in heterologous gene expression and co-culture dynamics are evident, BC modification through genetic engineering and co-culturing strategies remains achievable across all tested Komagataeibacter species. Our work provides a comparative framework to guide researchers in selecting optimal species based on their specific application requirements.

|
Scooped by
mhryu@live.com
January 13, 6:30 PM
|
The Antarctic ecosystems are a combination of conditions, including extremely low values of temperature and humidity. Nonetheless, some organisms, such as fungi, can adapt to these extreme conditions. The environmental temperature is one of the parameters thoroughly affecting the structure and composition of fungal membrane lipids. The psychrophilic fungi generally increase the disorder within macromolecules to maintain membrane fluidity at low temperatures. To do so, Antarctic fungi increase the proportion of unsaturated fatty acids that allow maintaining a semi-fluid state of the membranes and survive at extremely low temperatures. This ecological feature may be of interest for the characterization of phenotypical traits of the ecological adaptation of these fungi to the extreme environmental conditions of Antarctica. Moreover, this can be of inspiration to find solutions inspired by nature for alternative sources of polyunsaturated fatty acids (PUFAs) for diets of humans and animals. We characterized three fungal strains isolated from Antarctica and set up a laboratory/small-scale production of fungal biomass with a high content of beneficial PUFAs. In detail, three fungal species previously isolated from Antarctic environmental matrices were tested and identified at the genome level. We also conducted growth experiments to determine the effects of temperature and substrate on biomass and PUFA production. The results showed that these fungi have a high percentage of PUFAs compared to saturated ones; the growth at low temperatures (10°C) increases the production of linolenic acid (C18:3) while the biomass amount (yield) depends on the composition of the growth substrate; a satisfying qualitative-quantitative production is achieved using agri-food chain waste products, such as brewing and whey waste, as a growth substrate.

|
Scooped by
mhryu@live.com
January 13, 6:02 PM
|
Global climate change poses increasing threats to seed production and thus food security. The seed microbiome plays an essential role in regulating the whole seed life cycle. Specific seed endophytes and spermosphere microorganisms orchestrate the maintenance and termination of dormancy towards the synchronization of germination plasticity to meet agricultural demands. In this review, we summarize recent advances by linking seed-microbiome interactions with seed processes. We review the sources of seed microbiomes and their physiological regulation on dormancy and germination in response to environmental changes with a focus on phytohormone crosstalk. We also discuss the molecular mechanisms by which seed-microbe interactions affect seed destiny. Finally, we explore emerging precision applications of microbiomes in the seed industry by integrating cutting-edge technologies such as microbial seed coatings and artificial intelligence (AI) in seed science and technology. In conclusion, harnessing microbiome-based strategies to manipulate seed life cycle holds immense promise for sustainable food production in a changing global climate.

|
Scooped by
mhryu@live.com
January 13, 4:21 PM
|
Advances in synthetic biology continue to potentiate bacterial cancer therapy. Here, we constructed biosensor-driven encapsulation systems for autonomous control of capsular polysaccharides of Escherichia coli Nissle 1917 to improve pharmacokinetic profiles. The engineered bacteria were programmed to express capsular polysaccharides for immune evasion upon intravenous administration to reach tumors and then turn off gene expression upon colonizing the tumors based on quorum-sensing or acid-sensing to prevent dissemination of bacteria into the systemic circulation. Because a classical pharmacokinetic model could not capture the dynamic nature of living therapeutics, a two-state pharmacokinetic model was developed to simulate the autonomous control of capsular polysaccharides in different biological compartments and their impact on biodistribution. Using this model, we identified parameters in gene circuit dynamics and immune clearance that influence tumor colonization and systemic bacterial persistence. In a “humanized” pharmacokinetic model with an increased rate of complement-mediated lysis of bacteria, biosensor-driven systems achieved tumor seeding densities comparable to wild-type bacteria while reducing bacterial loads in blood and liver by several orders of magnitude, highlighting their potential for safe systemic delivery. The biosensor-driven systems represent a more effective strategy to control living drugs than inducible systems, and the two-state pharmacokinetic model is a first step to capture the autonomous nature of this new class of therapeutics for clinical translation.

|
Scooped by
mhryu@live.com
January 13, 3:56 PM
|
Gene expression is typically studied on a gene-by-gene basis, with regulation analyzed primarily in response to environmental or developmental cues. In contrast, much less is known about how intrinsic factors, such as cell size or DNA content, influence global gene expression patterns. Cell size varies significantly across different cell types and dynamically changes during the cell cycle. To maintain proper intracellular concentrations of biomolecules such as mRNAs and proteins, gene expression must be coordinated with cell size. Emerging evidence from diverse organisms, including bacteria, yeast, animals, and plants, demonstrates that transcriptional output scales with cell size, suggesting a conserved principle of gene regulation. However, the mechanisms by which cells sense their size and modulate gene expression accordingly remain poorly understood. In this review, we summarize recent advances in uncovering the molecular and cellular principles of gene expression scaling with cell size across kingdoms. We also highlight key open questions in the field, with a particular emphasis on how plant systems, still underexplored in this context, can provide additional insights into the fundamental principles of size-dependent gene regulation.

|
Scooped by
mhryu@live.com
January 13, 3:41 PM
|
TnpB is a compact RNA-guided endonuclease and evolutionary ancestor of CRISPR-Cas12 that offers a promising platform for genome engineering. However, the genome-editing activity of TnpBs remains limited and its underlying determinants are poorly understood. Here, we used biochemical and single-molecule assays to examine the DNA-unwinding mechanism of Youngiibacter multivorans TnpB (Ymu1 TnpB). DNA unwinding proceeds through formation of a partially unwound intermediate state to a fully unwound open state. The open state forms inefficiently and collapses readily in the absence of negative supercoiling. An optimized variant, Ymu1-WFR, stabilizes formation of both the intermediate and open states, resulting in enhanced DNA cleavage in vitro and increased genome editing in vivo. These findings identify the physical basis for the observed minimal activities of natural TnpBs, revealing how stabilizing specific unwinding states enables efficient DNA targeting.

|
Scooped by
mhryu@live.com
January 13, 3:15 PM
|
Generative protein modeling provides advanced tools for designing diverse protein sequences and structures. However, accurately modeling the conformational landscape and designing sequences remain critical challenges: ensuring that the designed sequence reliably folds into the target structure as its most stable conformation, and optimizing the sequence for a given suboptimal fixed input structure. In this study, we present a systematic analysis of jointly optimizing sequence-to-structure and structure-to-sequence mappings. This approach enables us to find optimal solutions for modeling the conformational landscape. We validate our approach with large-scale protein stability measurements, demonstrating that joint optimization is superior for designing stable proteins using a joint model (TrRosetta and TrMRF) and for achieving high accuracy in stability prediction when jointly modeling (half-masked ESMFold pLDDT + ESM2 Pseudo-likelihood). We further investigate features of sequences generated from the joint model and find that they exhibit higher frequencies of hydrophilic interactions, which may help maintain both secondary structure registry and pairing-features not captured by structure-to-sequence modeling alone. This study presents a comprehensive modelling framework that jointly optimizes sequence and structure to generate de novo proteins with improved folding stability, providing large-scale experimental benchmarking across multiple computational design methods

|
Scooped by
mhryu@live.com
January 13, 2:22 PM
|
Translation is a fundamental process for every living organism. In plants, the rate of translation is tightly modulated during development and in responses to environmental cues. However, it is challenging to measure the actual translation state of the tissues in vivo. Here, we report the introduction of an in vivo translation marker based on bimolecular fluorescence complementation, the Ribo-BiFC. We combined a method originally developed for the fruitflies with an improved low background split-mVenus BiFC system previously described in plants. We labelled small subunit ribosomal proteins (RPS) and large subunit ribosomal proteins (RPL) of Arabidopsis thaliana with fragments of the mVenus fluorescent protein (FP). We tested the Ribo-BiFC method using transiently expressed recombinant ribosomal proteins in epidermal cells of Nicotiana benthamiana. The BiFC-tagged ribosomal proteins complemented the mVenus molecule and were detected by fluorescence microscopy, potentially visualizing the close proximity of translating assembled 80S ribosomal subunits. Although the resulting signal is less intense than that of known interactors, its detection points to the functionality of the system. This Ribo-BiFC approach has further potential for use in stable transgenic lines in enabling the visualization of translational rate in plant tissues and changing translation dynamics during plant development, under abiotic stress or in different genetic backgrounds.

|
Scooped by
mhryu@live.com
January 13, 1:12 PM
|
Nutrient crossfeeding critically governs microbiome–host interactions and ecosystem stability. Cobamides, synthesized only by prokaryotes, offer a powerful and tractable model for studying nutrient-mediated interdependencies in soil food webs; however, their ecological role in sustaining soil health remains unclear. Here, we construct the Soil Cobamide Producer database (SCP v.1.0) by integrating over 48,000 metagenomic and genomic datasets from 1,123 sampling sites. This database catalogs phylogenetically diverse prokaryotes (19 phyla, 302 genera) with cobamide biosynthetic potential. Using this resource, we identify host-specific colonization patterns of cobamide-producing microbes in fauna. These microbes also carry diverse functional traits that may contribute to trophic cascades and microbial community stability. In an Enchytraeid model, these colonizers support host development, modulate gene expression, and promote gut stability through transkingdom interactions, with cobamide biosynthesis serving as one representative trait among multiple microbial functions. At macroecological scales, cobamide-producing microbes occur across relatively high trophic levels, reflecting a broader principle of nutrient transfer that may also apply to other essential metabolites. This framework provides a general basis for studying nutritional microbes in soil food webs and advances One Health research. Soil life relies on microbial nutrient exchange to maintain ecosystem balance. This study shows that cobamide-producing microbes act as essential connectors in soil food webs, supporting host health and highlighting cross-kingdom links central to One Health.
|
 Your new post is loading...
Your new post is loading...



























1str