Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 4:55 PM
|
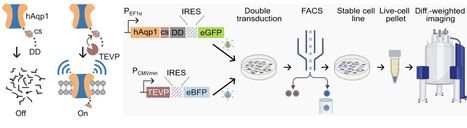
Creating genetic sensors for noninvasive visualization of biological activities in optically opaque tissues holds immense potential for basic research and the development of genetic and cell-based therapies. Magnetic resonance imaging (MRI) stands out among deep tissue imaging methods for its ability to generate high-resolution images without ionizing radiation. However, the adoption of MRI as a mainstream biomolecular technology has been hindered by the lack of adaptable methods to link molecular events with genetically encodable contrast. Here, we introduce modular aquaporin-based protease-activatable probes for enhanced reporting (MAPPER), a platform for the systematic creation of genetic sensors for MRI. To develop MAPPER, we engineered protease-activatable MRI reporters using two approaches: protein stabilization and subcellular trafficking. We established the applicability of MAPPER in distinct mammalian cell types and demonstrated its versatility by assembling genetic sensors for diverse targets without requiring extensive customization for each target. MAPPER provides a programmable platform for streamlining the development of noninvasive, nonionizing genetic sensors for biomedical research and in vivo diagnostics.

|
Scooped by
mhryu@live.com
Today, 3:43 PM
|
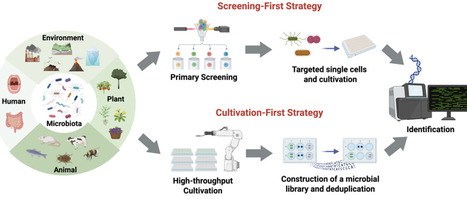
The growing demand for novel antibiotics, industrial enzymes, and environmentally sustainable biotechnological solutions is drawing increasing attention to more efficient strategies for microbial resource discovery across biomedical, industrial, and ecological domains. To meet this need, high-throughput microbial culturomics has emerged as a powerful strategy that integrates advanced cultivation platforms, diverse growth conditions, and rapid identification technologies. This review explores two principal paradigms within high-throughput microbial culturomics: the function-driven screening-first strategy and the enrichment-based cultivation-first strategy. Their technical foundations, application scenarios, and inherent limitations are examined in detail, providing a comparative analysis of their respective advantages and challenges. We further emphasize the importance of high-throughput identification methods, which play a crucial role in classifying isolates and revealing their functional potential. Ultimately, the review explores future directions for the development of automated, fully integrated cultivation platforms that integrate large-scale experimentation with data-driven optimization. By offering a structured comparison of culturomics strategies, integration pathways, and key obstacles, this review serves as a methodological reference for advancing microbial isolation, functional screening, and biotechnological innovation.

|
Scooped by
mhryu@live.com
Today, 2:43 PM
|
Most antibiotics are natural compounds or their derivatives, and bacteria have evolved defensive mechanisms to resist them. Many of these mechanisms are still poorly understood or unknown. This study reveals that in Bacillus subtilis, the transcription factor HelD increases resistance to rifampicin by protecting its target, RNA polymerase (RNAP). This protection is mediated by the HelD N-terminal domain that penetrates into RNAP to the close vicinity of the rifampicin binding pocket. Importantly, the bacterium detects low rifampicin levels using a unique regulatory system involving two convergent promoters with finely tuned kinetic properties. In the absence of rifampicin, the stronger antisense promoter inhibits transcription from the sense promoter. In the presence of subinhibitory rifampicin concentration, the antisense promoter is more likely to encounter rifampicin-bound RNAP. This relieves the repression from the sense promoter, increasing its transcription by almost two orders of magnitude, boosting helD expression. A similar two-promoter arrangement also controls the pps gene, which encodes a rifampicin-modifying enzyme. These findings define a widespread bacterial response system sensitive to rifampicin, as this dual-promoter architecture is conserved across many bacterial species and found upstream of genes potentially involved in rifampicin resistance, such as those for hydrolases, transporters, and transferases.

|
Scooped by
mhryu@live.com
Today, 1:49 PM
|
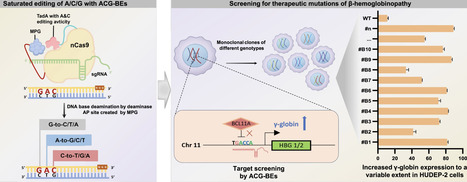
Current base editors act on a maximum of two base substrates and generate limited base conversions or transversions, hindering their applicability for inducing DNA sequence diversity. Here, we engineered a triple base editor (named ACG-BEs) using a fusion of adenine base editor with high A/C catalytic activity and evolved N-methylpurine DNA glycosylase. ACG-BEs enables efficient, multiplexed saturation mutagenesis across adenine (A), cytosine (C), and guanine (G), achieving conversion efficiencies of up to 80.5% for A-to-G/C/T, 75.8% for C-to-T/G/A, and 63.4% for G-to-C/T/A in HEK293T cells. Leveraging ACG-BEs, we identify novel mutations in the HBG1/2 promoter region that confer efficient activation of γ-globin expression in HUDEP-2 cells—a promising advancement for therapeutic strategies targeting hemoglobinopathies. These findings highlight ACG-BEs as a cutting-edge platform for multiplexed saturation mutagenesis, offering broad applications in genetic screening and therapeutic base mutation introduction through enhanced DNA sequence diversity.

|
Scooped by
mhryu@live.com
Today, 1:22 PM
|
RNAs play crucial roles in various important biological functions such as gene regulation and catalysis. The functions of RNAs are generally coupled to their structures as well as to the stability of the structure, which can strongly depend on ionic conditions. However, it is still a challenge to make reliable predictions for the structures and stability of RNAs in ion solutions. In this work, we developed a coarse-grained method involving temperature and ion effects for simulating RNA folding and three-dimensional (3D) structure prediction, named TiRNA. Extensive tests demonstrate that TiRNA can make successful predictions for 3D structures of RNAs, including pseudoknots and multi-way junctions, and for thermal stability of RNAs in ion solutions solely from sequences, as compared with the top existing methods. Moreover, TiRNA can also make reliable predictions for 3D structures and stability of RNAs in ion solutions based on inputting secondary structures.

|
Scooped by
mhryu@live.com
Today, 1:04 PM
|
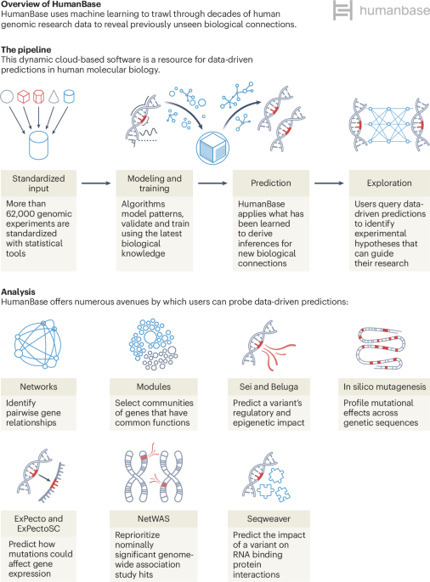
Increasing quantities of genomic data present an opportunity for uncovering functional and mechanistic insights. However, given the complexity and scale of the data, sophisticated analytical frameworks and integrative approaches are required. In addition, the unique modeling requirements of different data modalities and the large number of disease states, tissue types and developmental profiles pose complex challenges. Current analysis methods are often computationally expensive and technically challenging for the end user to apply. There is a need for comprehensive resources that bridge the gap between raw data and biological insights, allowing researchers at all levels of computational fluency to bring large data compendia and cutting-edge analytical approaches to bear on their questions of interest. Here, we present HumanBase (https://humanbase.io), an interactive web-based platform that uses AI to integrate and interpret large-scale human genomic data.

|
Scooped by
mhryu@live.com
Today, 10:56 AM
|
Here, we present an efficient PE system in rice, developed using a combination of engineered pegRNA (epegRNA) to enhance stability and prevent degradation of the 3′ extension, and paired pegRNAs to target different DNA strands, synthesizing edited DNA sequences simultaneously by RT of each template. Using this approach, we observed that undesired scaffold-derived mutations occurred more frequently in regenerated rice plants than indels resulting from DNA double-strand break (DSB) repair. To improve the fidelity of the precise edits, we next tested a PE system using paired epegRNA-HFs (PE with paired epegRNA-HFs). Our results show that PE with paired epegRNA-HFs in rice avoids the introduction of undesired scaffold-derived byproducts into the target locus, resulting in the establishment of an efficient and accurate PE system in plant cells.

|
Scooped by
mhryu@live.com
Today, 10:39 AM
|
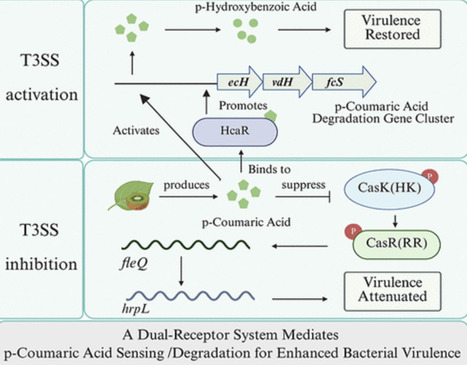
Plants deploy diverse secondary metabolites for chemical defense against pathogens, and in response, phytopathogens have evolved elaborate counterstrategies to subvert host immunity. In this study, we demonstrate that Pseudomonas syringae pv actinidiae strain M228 (Psa_M228)─the causal agent of kiwifruit bacterial canker (KBC)─employs a dual-functional system to sense and degrade host-derived p-coumaric acid (p-CA), thereby neutralizing the host’s chemical defense. Specifically, the kiwifruit host plant produces p-CA, which binds to histidine kinase CasK of Psa_M228’s two-component signaling system CasKR, inhibiting response regulator CasR phosphorylation. This reduces CasR binding to the promoter region of fleQ, downregulating hrpL expression─a master regulator of Type III secretion system (T3SS), ultimately attenuating virulence. For counter-defense, Psa_M228 utilizes the HcaR (hydroxycinnamic acid regulator) receptor and hca gene cluster (encoding p-CA-degrading enzymes) to catabolize p-CA. This adaptation helps Psa_M228 evade plant immunity and restore virulence, revealing a host–pathogen arms race.

|
Scooped by
mhryu@live.com
Today, 1:07 AM
|
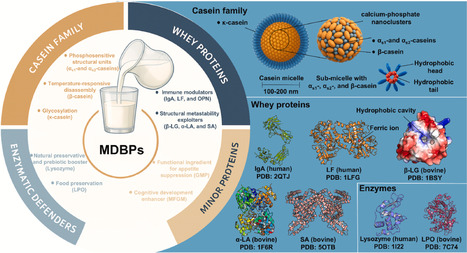
Milk-derived bioactive proteins (MDBPs) are essential for human growth, immune regulation, and neurodevelopment, possessing unique structure-function characteristics absent in plant-based and cultured proteins. Current industrial production adopts membrane separation, chromatography, and precipitation, but these methods face limited raw materials and high costs. Synthetic biology-enabled microbial cell factories (MCFs) offer a sustainable alternative, yet achieving both native-level bioactivity and high yield remains a substantial challenge. This review summarizes MDBPs from bovine and human milk, including their composition, abundance, structures, and physiological roles. It further delineates a complete recombinant production framework, encompassing chassis cell selection, transcriptional and translational optimization, post-translational modifications (PTMs), intracellular trafficking, secretion, and proteolytic stability. Particular attention is devoted to advanced microbial cell factory (MCF) strategies that integrate systems biology, multi-omics analytics, and protein engineering to address production bottlenecks, thereby providing a coherent technical roadmap for the industrial translation of MDBP research. MCFs provide a sustainable platform for MDBP manufacturing, offering high process controllability and scalability. Notably, achieving precise recapitulation of complex PTMs and optimizing secretion pathways are key to ensuring higher bioactivity and yield. Future artificial intelligence (AI)-assisted design-build-test-learn (DBTL) cycles will further improve MCF engineering and broaden MDBP applications in functional foods, clinical nutrition, and targeted therapeutics.

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
Bacterial chromosomes contain Genomic Islands carrying genes involved in virulence, mutualism, and resistance to antibiotics that are transferred horizontally. Some of them conjugate autonomously (Integrative Conjugative Elements, ICEs), while others use relaxases to hitch on the former's conjugation machinery (Integrative Mobilizable Elements, IMEs). Yet, the mobility mechanism of most Genomic Islands remains elusive. Here, we explore the hypothesis that many carry origins of transfer (oriTs) by conjugation. Since very few known oriTs were found in ICEs and IMEs, we identified 52 novel families of oriTs that cover most integrative elements in six major nosocomial species. Most of these oriTs are specific to integrative elements, suggesting a clear split in the targets of hitchers of conjugative elements between plasmids and integrative elements. Among 7,363 Genomic Islands, we discovered more than 1,500 IMEs carrying an oriT and lacking relaxases and MPF systems. These elements, coined iOriTs, form diverse large ancient families that can be found in different species. These hitchers are genetically unrelated to the putative helpers, beyond the similarity at the oriT sequence, and often outnumber them. They include well-known pathogenicity islands, for which the mechanisms of mobility remained unknown. Unlike plasmids, iOriTs have few antibiotic resistance genes, but high density of virulence factors. Like plasmids, the vicinity of oriTs concentrates defense and counter-defense systems potentially favoring Genomic Islands dissemination. Hence, iOriTs are frequent integrative mobile genetic elements that evolved to transfer horizontally by hitching on other elements while contributing to bacterial genetic adaptation.

|
Scooped by
mhryu@live.com
Today, 12:47 AM
|
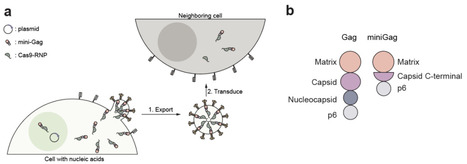
Genome editing enzymes have vast therapeutic potential. However, achieving sufficient delivery in vivo remains a major challenge, because editing machinery is confined to the subset of transfectable cells in a tissue. Here, we tested the possibility that genome editing could be amplified in vivo by programming transfected cells to produce and transfer editing enzymes in lipid vesicles to neighboring cells. Our data show that this NANoparticle-Induced Transfer of Enzyme (NANITE) strategy tripled editing efficiency in cultured cells relative to non-spreading controls. Furthermore, a single intravenous injection of the NANITE plasmid into mice induced ~3-fold higher levels of liver editing at the Ttr locus relative to non-spreading controls, with corresponding reductions in serum transthyretin levels. Amplifying therapeutic enzymes in situ offers a nonviral and non-infectious strategy to overcome low delivery efficiencies and reduce effective dose requirements.

|
Scooped by
mhryu@live.com
January 13, 11:59 PM
|
Bacterial cellulose (BC) is produced by diverse bacterial species, including members of the genus Komagataeibacter, and has emerged as a potential sustainable alternative to conventional materials such as plastics and leather. Its production by bacteria is rapid and easily scalable, and via genetic modifications or culturing the bacteria with other microbes, it is possible to rationally enhance the BC that is produced, for example by adding functional components. Here, we systematically evaluated BC functionalization strategies, including genetic engineering and co-culturing with other engineered microbes across the most widely used Komagataeibacter species: K. rhaeticus, K. xylinus, K. medellinensis and K. sucrofermentans. We established that all tested strains are amenable to DNA transformation and capable of expressing heterologous genes from identical genetic constructs. However, we identified species-specific differences in heterologous gene expression, cellulose production in varying environmental conditions and in the abilities of the strains to co-culture with Escherichia coli. Additionally, we demonstrated that all these cellulose-producing bacteria can establish functional symbiotic co-cultures with yeast. While inter-species variations in heterologous gene expression and co-culture dynamics are evident, BC modification through genetic engineering and co-culturing strategies remains achievable across all tested Komagataeibacter species. Our work provides a comparative framework to guide researchers in selecting optimal species based on their specific application requirements.

|
Scooped by
mhryu@live.com
January 13, 6:30 PM
|
The Antarctic ecosystems are a combination of conditions, including extremely low values of temperature and humidity. Nonetheless, some organisms, such as fungi, can adapt to these extreme conditions. The environmental temperature is one of the parameters thoroughly affecting the structure and composition of fungal membrane lipids. The psychrophilic fungi generally increase the disorder within macromolecules to maintain membrane fluidity at low temperatures. To do so, Antarctic fungi increase the proportion of unsaturated fatty acids that allow maintaining a semi-fluid state of the membranes and survive at extremely low temperatures. This ecological feature may be of interest for the characterization of phenotypical traits of the ecological adaptation of these fungi to the extreme environmental conditions of Antarctica. Moreover, this can be of inspiration to find solutions inspired by nature for alternative sources of polyunsaturated fatty acids (PUFAs) for diets of humans and animals. We characterized three fungal strains isolated from Antarctica and set up a laboratory/small-scale production of fungal biomass with a high content of beneficial PUFAs. In detail, three fungal species previously isolated from Antarctic environmental matrices were tested and identified at the genome level. We also conducted growth experiments to determine the effects of temperature and substrate on biomass and PUFA production. The results showed that these fungi have a high percentage of PUFAs compared to saturated ones; the growth at low temperatures (10°C) increases the production of linolenic acid (C18:3) while the biomass amount (yield) depends on the composition of the growth substrate; a satisfying qualitative-quantitative production is achieved using agri-food chain waste products, such as brewing and whey waste, as a growth substrate.
|

|
Scooped by
mhryu@live.com
Today, 4:47 PM
|
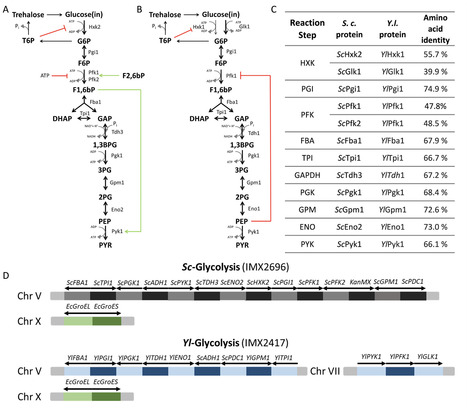
Protein allostery, present in all three domains of life, is key to the regulation of metabolism by allowing fast and precise control of catalysis in response to cellular demands. While metabolic pathways are frequently equipped with multiple allosterically regulated catalytic steps, experimental studies often focus on a single step, failing to capture how regulations exerted at multiple steps interact with each other for tuning pathways. Using the nearly ubiquitous Embden-Meyerhof-Parnas pathway of glycolysis as a paradigm, the present study unveils a remarkable regulatory synergy between multiple allosteric proteins of a metabolic pathway and demonstrates its impact on cell survival in dynamic environments. By using complete pathway complementation, as well as single-gene complementation, the essential regulatory steps were identified to be glucokinase, phosphofructokinase, and pyruvate kinase. Expression of these enzymes together, even in the context of the Saccharomyces cerevisiae pathway, led to imbalances in glycolysis that could only be overcome by lowering the glucokinase activity. Integrating these results with kinetic modeling and microfluidics experiments, the present work reveals the key synergistic role played by allosteric regulations in preventing glycolytic imbalance in the model eukaryote Saccharomyces cerevisiae and highlights the power of synthetic biology in addressing long-standing questions in systems biology.

|
Scooped by
mhryu@live.com
Today, 3:19 PM
|
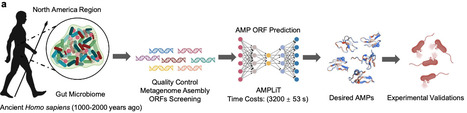
Fecal coprolites preserve ancient microbiomes and are a potential source of extinct but highly efficacious antimicrobial peptides (AMPs). Here, we develop AMPLiT (AMP Lightweight Identification Tool), an efficient tool deployable to portable hardware for AMP screening in metagenomic datasets. AMPLiT demonstrates AUPRC performances of 0.9486 ± 0.0003 and reasonable overall training time of 3200 ± 53 s. By computationally utilizing AMPLiT, we analyze seven ancient human coprolite metagenomes, identifying 160 AMP candidates. Of 40 representative peptides synthesized, 36 (90%) peptides demonstrate measurable antimicrobial activity at 100 μM or less in vitro. Strikingly, approximately two-thirds of these peptides are sourced from Segatella copri, a dominant ancient gut commensal that is conspicuously underrepresented in modern populations, particularly those with Westernized lifestyles. Representative S. copri-derived AMPs exhibit disruptions against membranes of pathogenic bacteria, coupled with low cytotoxicity and hemolytic risk. In vivo, lead peptides demonstrate potent antibacterial and wound-healing efficacy comparable to traditional antibiotics, especially in combating gram-positive pathogens. Our findings highlight the ancient gut microbiomes as sources of novel AMPs, offering valuable insights into the historical role of S. copri in human health and its decline in contemporary populations. Here, the authors develop AMPLiT a tool for screening antimicrobial peptides in metagenomic datasets, and apply it to human coprolite metagenomes, finding that Segatella copri, an ancient prevalent human gut bacterium declined in modern populations, harbors unexplored antimicrobial reservoir, offering an alternative approach against modern pathogenic infections.

|
Scooped by
mhryu@live.com
Today, 1:56 PM
|
CRISPR-Cas12a holds substantial promise for molecular diagnostics, yet its rapid and uncontrolled activation often results in background leakage and disrupts the coordination of upstream reaction modules. Here, we established a steric-regulation framework that enables predictable tuning of Cas12a trans-cleavage kinetics through rationally engineered extensions on split activators. Systematic analysis of extension orientation, length, and hybridization state revealed quantitative and direction-dependent rules governing steric control of activator assembly and Cas12a activation. Guided by these insights, we integrated the sterically regulated split activator into an entropy-driven DNA circuit to construct a fully one-pot cascaded detection system. The engineered steric barriers effectively suppressed premature activation and established precise kinetic matching between the DNA circuit and Cas12a. The resulting platform achieved a detection limit of 1.24 pM for microRNA-21 and demonstrated high fidelity. This work defines a predictable steric-gating mechanism for Cas12a activation and delivers a nucleic-acid-only regulatory module that can be incorporated into diverse CRISPR architectures, supporting the development of robust, leakage-resistant one-pot diagnostic systems.

|
Scooped by
mhryu@live.com
Today, 1:42 PM
|
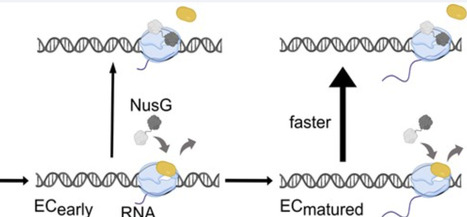
The E. coli transcription initiation factor σ70 was long believed to dissociate from RNA polymerase after promoter escape, following synthesis of short ~9–11 nt RNAs. However, recent evidence indicates σ70 is retained in elongation complexes (ECs), where it enhances pausing—implying that stochastic or factor-mediated displacement is required for processive elongation. Using fluorescence correlation spectroscopy, we quantified σ70 retention in the presence of elongation factors NusG and NusA, demonstrating that NusG—but not NusA—actively promotes σ70 dissociation. Single-molecule TIRF further revealed that NusG-mediated σ70 dissociation becomes favorable as ECs progress, with lower energy barriers for displacement in later elongation stages. These findings suggest a broader mechanistic paradigm in which elongation factors actively displace initiation factors to regulate transcription—a process that may be conserved across all domains of life.

|
Scooped by
mhryu@live.com
Today, 1:14 PM
|
Enzymes are powerful and sustainable catalysts, but their widespread application is limited by the difficulty of identifying functional starting points for optimization, creating a major bottleneck in early- stage biocatalyst discovery. Designing libraries of such starting enzymes remains particularly challenging. Here, we use the GenSLM protein language model to generate novel β-subunit of tryptophan synthase (TrpB) enzymes that express in Escherichia coli and are both stable and catalytically active. Many generated TrpBs also display significant substrate promiscuity, outperforming their natural counterparts on non-native substrates. Some even surpass laboratory-evolved TrpBs. Comparison of the most-active and most-promiscuous generated TrpB to its closest natural homolog confirms that the enhanced versatility is absent from the natural enzyme, highlighting the creative potential of generative models. These results demonstrate that the generated TrpBs not only preserve natural structure and function but also acquire non-natural properties, establishing generative models as powerful tools for biocatalyst discovery and engineering. Enzymes are highly selective and sustainable catalysts for chemical synthesis, but their optimization is often limited by the difficulty of identifying functional starting points. This study shows that using the GenSLM protein language model to design TrpB variants can yield stable, active enzymes with broad substrate promiscuity, outperforming natural and evolved counterparts and demonstrating the potential of generative models to accelerate biocatalyst discovery.

|
Scooped by
mhryu@live.com
Today, 11:40 AM
|
Pseudomonas spp. are ubiquitous, metabolically versatile bacteria that exhibit remarkable adaptability to thrive in diverse ecological niches contaminated with aromatics, such as pesticide-polluted agricultural soils, industrial wastewater, and lignin-based waste. This review highlights the unique genetic and metabolic traits of Pseudomonas bharatica CSV86T, a novel soil bacterium capable of degrading a wide range of aromatics, including lignin-derived phenylpropanoid compounds. Unlike other pseudomonads, aromatic metabolism in strain CSV86T is not subjected to carbon catabolite repression by simple carbon sources. Instead, it preferentially utilizes aromatics over glucose/glycerol and co-metabolizes them with organic acids, circumventing a major bottleneck in biodegradation. The strain is plasmid-free and naphthalene metabolism pathway genes are present on conjugatively transferable integrative conjugative element, ICEnahCSV86, offering potential for genetic bio-augmentation. Notably, the strain grows slowly on glucose and metabolizes it exclusively via phosphorylative pathway, as oxidative routes are absent. Beyond aromatic metabolism, it displays multifarious plant growth-promoting and beneficial eco-physiological traits for niche colonization and adaptation, crucial for restoration of contaminated sites. Collectively, these unique traits position strain CSV86T as a niche-adapted alternative to model biodegradation strains, such as P. putida KT2440. Its potential can be further leveraged through metabolic engineering for detection, degradation, and up-cycling of aromatic pollutants.

|
Scooped by
mhryu@live.com
Today, 10:48 AM
|
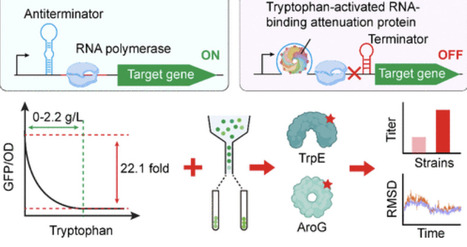
Biosensors have been widely applied for high-throughput strain screening and dynamic regulation of metabolic networks. However, existing tryptophan sensors based on transcription factors or riboswitches often suffer from a narrow dynamic range and limited response threshold. In this study, we developed a series of tryptophan-responsive biosensors in E. coli using the tryptophan-activated RNA-binding attenuation protein (TRAP) as the sensing module. First, we validated TRAP functionality and engineered a functional biosensor by fine-tuning its expression. Subsequently, screening of TRAP variants and optimization of TRAP–leader sequence interactions yielded two biosensors that exhibited distinct dynamic ranges (up to 22.1-fold) and response thresholds of 0–2.2 g/L, respectively. Using these biosensors, we screened two beneficial variants of key rate-limiting enzymes in the tryptophan biosynthetic pathway and further investigated their catalytic mechanisms through molecular dynamics simulations. Collectively, this study provides tools for engineering high tryptophan-producing strains and new strategies for biosensor design.

|
Scooped by
mhryu@live.com
Today, 1:10 AM
|
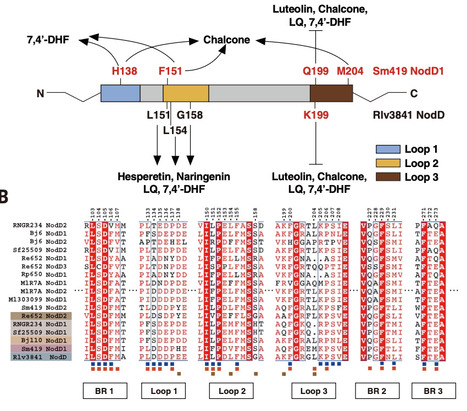
The specific partnership between legumes and rhizobia relies on a chemical dialogue. Plant flavonoids activate the bacterial transcription factor NodD, which triggers production of Nod factors that are recognized by the plant. Structural studies of the Pisum sativum (pea) symbiont Rhizobium leguminosarum NodD revealed two pockets that are essential for its activation by flavonoids. Comparative studies with NodD1 of Sinorhizobium medicae, the symbiont of Medicago truncatula, revealed that this specificity is determined by the shape of the pocket and by specific amino acids. A chimeric NodD containing the flavonoid recognition residues from S. medicae NodD1 in the R. leguminosarum NodD backbone was sufficient to complement nitrogen fixation in M. truncatula by an S. medicae nodD1 mutant, confirming the critical role of flavonoid recognition in host range.

|
Scooped by
mhryu@live.com
Today, 1:04 AM
|
Phosphorus (P) is a primary mineral nutrient essential for the growth and productivity of many crop plants. Although abundant in nature, its bioavailability is limited due to the prevalence of insoluble forms. An alternative for meeting agricultural P demand is the application of P-solubilizing microorganisms (PSMs), which mobilize it. Although progress has been made in the study of PSMs, knowledge gaps still exist regarding their role in sustainable agriculture. Therefore, this review examines the barriers to P acquisition in low-solubility soils and highlights recent advances in understanding the mechanisms of P solubilization mediated by plant-associated bacteria and fungi. The molecular strategies involved in the uptake and transport of P from soil in plants are also analyzed. Bacteria from genera such as Bacillus, Pseudomonas, and Streptomyces, as well as fungi including arbuscular mycorrhizal fungi, Aspergillus spp., and Penicillium spp., employ various approaches to solubilize P, leading to improved plant nutrition. These mechanisms, which include the production of organic acids, cation chelation, proton exudation, and phosphatase activity, can be inferred from experimental approaches or genome mining strategies. The role of PSMs as plant growth promoters and enhancers of plant nutrition across diverse environmental conditions are also discussed. Finally, we propose the integration of PSM consortia as multifunctional bioinoculants to promote sustainable agricultural practices.

|
Scooped by
mhryu@live.com
Today, 12:49 AM
|
The alpha-proteobacterium Zymomonas mobilis exhibits exceptional ethanologenic physiology, which makes it a traditional alcoholic beverage producer and a promising chassis for biofuel production. Although genetic tools for this organism have expanded in recent years, a fundamental aspect of its chromosome organization remains to be understood. In particular, Z. mobilis has been suggested to exhibit polyploidy, but this feature is not fully confirmed because of discrepancies among studies reporting the copy number of chromosomes. Here, we tagged the chromosome-partitioning protein ParB with a fluorescent marker to visualise its cellular localisation and estimate chromosome copy number in individual cells. Imaging showed that Z. mobilis exhibits several distinctive ParB foci throughout the cytoplasm and an accumulated focus at the pole, demonstrating that a single Z. mobilis cell contains at >5 copies of the chromosome at the oriC regions. After verifying its polyploidy, we sought to establish an efficient counter-selection system, which is crucial for engineering multiple copies of the chromosome. We assessed the efficacy of levan-sucrase (SacB) toxicity in Z. mobilis. We found that, despite Z. mobilis secreting a native extra-cellular sucrase SacB, heterologous periplasmically-localised Bacillus subtilis SacB rendered Z. mobilis cells sensitive to sucrose. We successfully used this effect for counter-selection when deleting and inserting targeted DNA sequences into the Z. mobilis genome. Together, this work provides important insights and tools for advancing Z. mobilis genetics and its biotechnological applications.

|
Scooped by
mhryu@live.com
Today, 12:42 AM
|
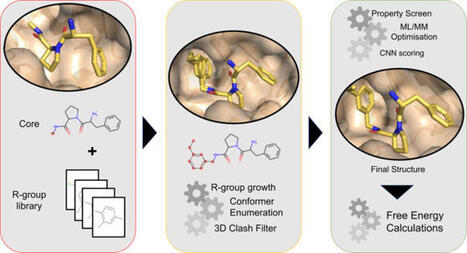
Automated free energy calculations for the prediction of binding free energies of congeneric series of ligands to a protein target are growing in popularity, but building reliable initial binding poses for the ligands is challenging. Here, we introduce the open-source FEgrow workflow for building user-defined congeneric series of ligands in protein binding pockets for input to free energy calculations. For a given ligand core and receptor structure, FEgrow enumerates and optimizes the bioactive conformations of the grown functional group(s), making use of hybrid machine learning/molecular mechanics potential energy functions where possible. Low energy structures are optionally scored using the gnina convolutional neural network scoring function, and output for more rigorous protein–ligand binding free energy predictions. We illustrate use of the workflow by building and scoring binding poses for ten congeneric series of ligands bound to targets from a standard, high quality dataset of protein–ligand complexes. Furthermore, we build a set of 13 inhibitors of the SARS-CoV-2 main protease from the literature, and use free energy calculations to retrospectively compute their relative binding free energies. FEgrow is freely available at https://github.com/cole-group/FEgrow , along with a tutorial. Automated free energy calculations for the prediction of binding free energies of ligands to a protein target are gaining importance for drug discovery, but building reliable initial binding poses for the ligands is challenging. Here, the authors introduce an open-source workflow for building user-defined congeneric series of ligands in protein binding pockets for input to free energy calculations.

|
Scooped by
mhryu@live.com
January 13, 6:31 PM
|
Optimization of flux distribution in central carbon metabolism is important to improve the microbial productivity. As the number of precursors required for synthesis differs for each target compound, optimal flux distribution also varies. A library of mutant strains with diverse flux distributions can aid in optimal strain screening. Therefore, in this study, we aimed to construct a library of Escherichia coli strains with stepwise changes in flux distribution by introducing mutations into the ribosome-binding sites of key enzyme genes on its chromosome. We focused on the flux ratios at the glucose-6-phosphate and acetyl-CoA branch points to enhance mevalonate production. Mutations were introduced into the RBS of pgi and gltA to vary the flux ratios of the two pathway branches. Furthermore, a combinatorial repression library comprising 16 strains was constructed by varying pgi and gltA expression at four levels, and a plasmid containing mevalonate synthesis genes was introduced into each strain. Batch cultures were performed to obtain strains with mevalonate titers and yields 2.4- and 3.4-fold higher than those of the parent strain. Overall, our combinatorial suppression library of pgi and gltA facilitated the effective identification of mutants with optimal metabolism for mevalonate production.
|
 Your new post is loading...
Your new post is loading...