Your new post is loading...

|
Scooped by
?
Today, 11:24 AM
|
Microorganism culturing is essential in microbiological research, with the selection of suitable culture media being critical for successful microbial growth. Traditionally, this selection has relied on empirical knowledge or trial and error, often resulting in inefficiency. In this study, we analysed nutrient compositions from the MediaDive database to construct a dataset of 2369 media types. Leveraging this dataset and microbial 16S rRNA sequences, we developed 45 binary classification models using the XGBoost algorithm. These models demonstrated strong predictive performance, achieving accuracies ranging from 76% to 99.3%, with the top-performing models for J386, J50 and J66 media reaching 99.3%, 98.9% and 98.8%, respectively. The models effectively predicted growth conditions for various human gut microbes, confirming their practical utility. This research improves the efficiency of microbial cultivation and highlights the potential of machine learning to optimise culture media selection and advance microbiological studies.

|
Scooped by
?
Today, 11:13 AM
|
The genomic signatures of microbial domestication remain poorly understood within the context of natural population variation. Here, we demonstrate that Aspergillus oryzae, the filamentous fungus used in soy sauce production, shares more recent ancestry with a predominantly northern, largely non-aflatoxigenic population of Aspergillus flavus (population C). Strikingly, A. oryzae isolates also overlap with the recently described, clinically enriched A. flavus population D, suggesting the possibility of multiple domestication events. Although A. oryzae exhibits reduced virulence compared to A. flavus, all isolates tested retained pathogenicity in a zebrafish infection model. At the transcriptomic level, Aspergillus populations are significantly differentiated, with distinct responses to population density, indicating that population-specific transcriptomes adapt differently to ecological conditions. These differences extend beyond gene content and are not always explained by phylogenetic relationships, suggesting that phenotypic diversification occurs through the rapid reorganization of transcriptomic architectures. For example, A. oryzae displays significantly elevated expression of a module enriched for carbohydrate metabolism. Population-specific variation is also evident among secondary metabolite (SM) gene clusters. While A. oryzae shows markedly reduced expression of specific SM genes, particularly those involved in aflatoxin biosynthesis, this trend does not extend across the entire secondary metabolome. Using machine-learning-based gene regulatory network inference, we identified population-specific transcriptomic differences linked to distinct transcription factors, with evidence for both cis- and trans-acting regulatory divergence, but no changes in global regulators such as laeA. Together, these findings provide new insights into the domestication of A. oryzae, its global significance, and the microevolution of fungal secondary metabolic pathways.

|
Scooped by
?
Today, 10:29 AM
|
As the ability to synthesize complete genomes becomes increasingly accessible, the question of what should compose those sequences is becoming more prevalent. However, identifying genes essential for the survival of an organism is challenging, as gene essentiality is a nuanced concept that heavily depends on context. In this study, we identified growth medium-specific important genes by performing transposon mutagenesis in Escherichia coli BW25113 and sequencing mutant populations at multiple time points in three growth media. Our analysis revealed a core set of 412 important genes shared across all conditions, along with distinct medium-specific gene sets of varying sizes. By analyzing temporal variations in read counts for each gene, we identified additional sets of genes whose inactivation causes a lighter impact on fitness. We used this dataset to define medium-specific gene modules required to sustain robust growth under each condition. Our study underscores the context-dependent nature of gene essentiality and marks a step toward refining the concept from a universal list to a more nuanced, condition-specific framework, which will be invaluable for future genome design efforts.

|
Scooped by
?
October 3, 7:34 PM
|
Plasmids are a foundational research reagent and a key material in biopharmaceutical manufacturing. Plasmids require a backbone for propagation in E. coli, which typically contains an antibiotic resistance gene alongside a replication origin. As plasmids increasingly enter clinical applications, concerns are raised on the safety risks of antibiotic resistance genes. Additionally, these protein-coding genes occupy long stretches of DNA that incur significant metabolic burdens on host cells, which negatively impacts plasmid manufacturability and functionality, leading to high production cost and compromised clinical efficacy. Here, we describe miniVec, a novel miniaturized plasmid backbone devoid of protein-coding sequence, and instead expresses a small RNA to provide constant selective pressure capable of sustaining high plasmid copy numbers in plain culture media devoid of antibiotics or other chemical additives. This simplifies large-scale fermentation and greatly increases plasmid yield. Notably, miniVec confers enhanced functionality in a variety of applications such as chemical transfection, electroporation, virus packaging, transposon- or CRISPR-mediated genome integration, and in vivo naked DNA transfection and vaccination, while exhibiting no detectable immunogenicity or toxicity. These advantages establish miniVec as the next-generation plasmid platform for clinical applications, featuring improved safety that aligns with regulatory expectations, enhanced manufacturability leading to much higher yield and dramatic cost reduction, and augmented functionality in diverse applications.

|
Scooped by
?
October 3, 7:22 PM
|
The identification of complex spatial patterns of microbial communities in relation to their ecological niches is fundamental to understanding the mechanisms of ecological interactions among diverse organisms. This study introduces a novel three-dimensional (3D) sampling approach to examine the spatial dynamics of microbial populations and niche differentiation influenced by plant-mediated effects in natural ecosystem. Microbial communities across horizontal and vertical dimensions were systematically mapped, and we found that the total microbial diversity, particularly among eukaryotes, increased more than ten-fold compared to that obtained via single-grid sampling, emphasizing the importance of spatial heterogeneity in shaping microbial dynamics. Moreover, the 3D framework enabled us to identify taxa specifically associated with particular plants, offering insights into plant–microbe interactions, pathogen prevalence, and ecological consequences of plant-driven effects on local communities. Collectively, these findings demonstrate that 3D sampling approach provides a reproducible and scalable methodology for investigating microbial spatial heterogeneity, pathogen ecology, and niche differentiation in natural environments.

|
Scooped by
?
October 3, 7:12 PM
|
Flavonoid O-glycosylation, catalyzed by uridine diphosphate (UDP)–glycosyltransferases, is crucial for their therapeutic efficacy. However, most UDP-glycosyltransferases encounter three major limitations: low activity, poor regioselectivity, and restricted substrate availability, hindering their pharmaceutical applications. To address these challenges, we conducted protein engineering on a previously unidentified glycosyltransferase, UGT75AJ2, which had 3′,7-O-glycosylation capabilities. Our approach involved three strategies: (i) development of a tailored focused rational iterative site-specific mutagenesis strategy, augmented by virtual screening and iterative mutagenesis, to design mutant Mut4-1 (S367A/V274A/F82V/I132T) with a 128-fold enhancement in relative catalytic activity; (ii) enhancement of the enzyme’s compatibility with a broader spectrum of sugar donors achieved through structural-based engineering, yielding mutant S14G/F366H/S367G and demonstrating effective utilization of diverse donors; (iii) construction of a targeted mutant library to enhance regioselectivity by active site analysis, leading to mutants with high selectivity for targeted glycosylation sites. This comprehensive study tackles predominant challenges in UDP-glycosyltransferases protein engineering, providing innovative approaches and insights that enhance the development of flavonoid glycosides.

|
Scooped by
?
October 3, 6:54 PM
|
Targeted degradation is emerging as a new therapeutic approach in the treatment of different diseases. It allows hijacking the cellular pathways deputed to protein or nucleic acid homeostasis to degrade a target macromolecule of interest involved in a pathogenic process. In the last decades, targeted protein degradation has been widely applied for the treatment of cancer or neurodegenerative disorders and some of such therapies are already in clinical use. More recently, therapeutic degraders such as PROTACs, LYTACs, HyTs, BacPROTACs, and others have also been explored in the field of antimicrobial and antiviral drug discovery. The peculiar mechanism of action, along with the opportunity to degrade both microbial and host targets, holds great promise for overcoming some limitations of classic antimicrobials, e.g. drug resistance, as well as for increasing the potency of current therapies. With a focus on the antimicrobial field, this Review aims at providing a comprehensive, state-of-the-art description of targeted degradation mechanisms and strategies developed so far, as well as to discuss advantages, disadvantages, and caveats of this innovative approach for combating infectious diseases.

|
Scooped by
?
October 3, 1:23 PM
|
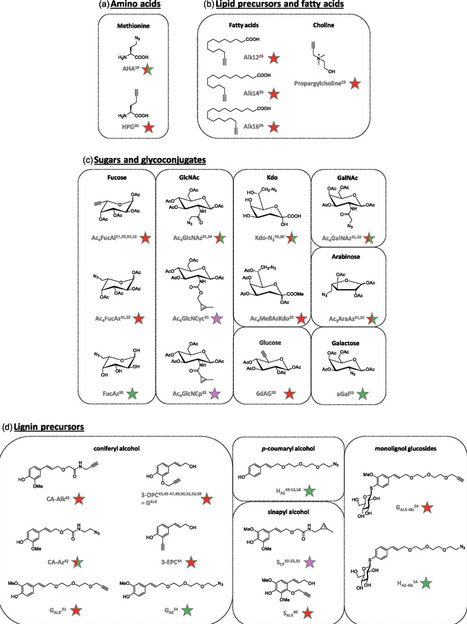
Chemical biology has reshaped the ability to investigate complex biological systems at the molecular level. In this context, chemical reporters have become important tools for labeling and tracking biomolecules in living systems with spatial and temporal precision. In plant biology, they provide an alternative to genetic approaches and allow the study of dynamic processes in species or organs that are not easily accessible. Through the use of click and bioorthogonal chemistry, small-molecule probes can be metabolically incorporated into specific molecular scaffolds such as sugars, monolignols, amino acids, and lipids. These probes make it possible to follow events like glycosylation, lignification, lipid turnover, or protein synthesis in living plant tissues. This review presents an overview of current chemical reporter strategies, from molecular design and synthetic considerations to their application in plant imaging. Herein, how these tools have contributed to the development of plant chemical biology by enabling precise and modular investigations of plant structure and metabolism is described. Herein, it is also examined how chemical reporters have entered interdisciplinary contexts, including collaborations between science and the arts. By converting molecular-level information into visual and sensory formats, these approaches open new perspectives for research, education, and communication across scientific and creative disciplines.

|
Scooped by
?
October 3, 10:53 AM
|
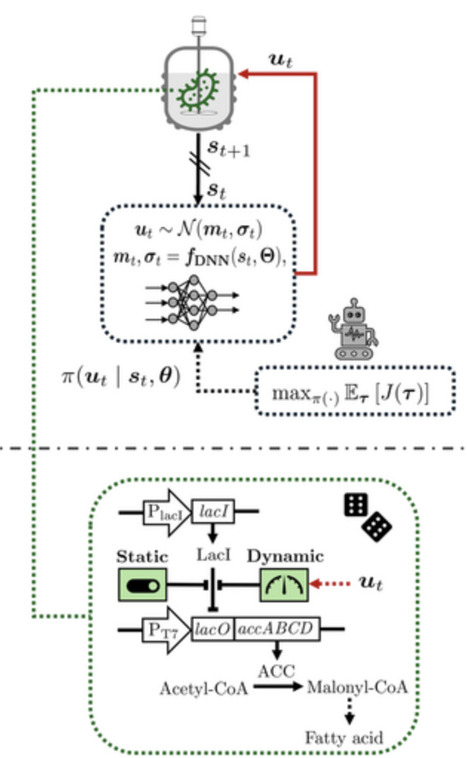
Dynamic metabolic control allows key metabolic fluxes to be modulated in real time, enhancing bioprocess flexibility and expanding available optimization degrees of freedom. This is achieved, for example, via targeted modulation of metabolic enzyme expression. However, identifying optimal dynamic control policies is challenging due to the generally high-dimensional solution space and the need to manage metabolic burden and cytotoxic effects arising from inducible enzyme expression. The task is further complicated by stochastic dynamics, which reduce bioprocess reproducibility. We propose a reinforcement learning framework to derive optimal policies by allowing an agent (the controller) to interact with a surrogate dynamic model. To promote robustness, we apply domain randomization, enabling the controller to generalize across uncertainties. When transferred to an experimental system, the agent can in principle continue fine-tuning the policy. Our framework provides an alternative to conventional model-based control such as model predictive control, which requires model differentiation with respect to decision variables; often impractical for complex stochastic, nonlinear, stiff, and piecewise-defined dynamics. In contrast, our approach relies on forward integration of the model, thereby simplifying the task. We demonstrate the framework in two Escherichia coli bioprocesses: dynamic control of acetyl-CoA carboxylase for fatty-acid synthesis and of adenosine triphosphatase for lactate synthesis.

|
Scooped by
?
October 3, 10:36 AM
|
Aiming to find novel ways to inhibit bacterial growth, we tested hammerhead ribozymes targeting the mRNAacpPtranscript, which encodes the essential acyl carrier protein in Escherichia coli. We engineered ribozymes with varying catalytic cores and arm lengths, finding that while short-armed ribozymes showed higher activity in vitro, long-armed variants demonstrated superior growth inhibition in vivo. Isothermal titration calorimetry confirmed tight binding between the ribozymes and the mRNA substrate, with association constants between 107 and 108 M−1, and gel electrophoresis verified substrate cleavage. Ribozymes were incorporated into bacterial plasmids, introduced via transformation into E. coli, and were expressed in a controlled manner, inhibiting bacterial growth by up to 70% over 24 h. Notably, ribozymes embedded within tRNA structures, a strategy intended to protect them from intracellular degradation, showed differential effectiveness compared to standalone variants; tRNA scaffolding preserved activity in long-armed but abolished it in short-armed constructs. Growth inhibition resulted from both mRNA cleavage and translational blocking, as demonstrated by comparing active ribozymes with their catalytically inactive variants. Furthermore, tetracycline efficacy was enhanced 2- to 4-fold in cells expressing ribozymes, indicating potential for synergy. This study demonstrates the first successful targeting of an essential gene in E. coli using hammerhead ribozymes, achieving growth inhibition through combined mechanisms of mRNA blocking and cleavage, and highlighting the potential of ribozymes as antibacterial strategies.

|
Scooped by
?
October 3, 10:28 AM
|
Bacteria exhibiting antimicrobial resistance (AMR) is a problem that has grown to become a significant global public health challenge. Antibiotic resistance genes (ARGs), determinants of AMR, mostly emerge from non-clinical settings. Identifying novel ARGs from the human microbiome which confer resistance to clinical antibiotics is a crucial component of addressing AMR. We sought to leverage the increased accuracy of computational protein structure prediction by training a one-class support vector machine on the pairwise primary protein structure and tertiary protein structure distributions of conserved structural regions of various ARG classes. We applied the computational method to seven human microbiome project reference strains and functionally confirmed eight out of nine tested novel ARG predictions, belonging to ARG classes: DFR, class B and C β-lactamases and penicillin binding proteins. We also applied the method directly to protein structures within the AlphaFold database and functionally confirmed a novel metallo-β-lactamase with low homology to existing ARGs. In total, 70% of tested genes confer resistance to clinical antibiotics at CLSI resistant breakpoints, or are emerging from the pre-resistome and may become clinically relevant in the future. This study represents a precise and computationally efficient method of identifying previously uncharacterized ARGs from DNA databases.

|
Scooped by
?
October 3, 10:14 AM
|
Engineering Non-Ribosomal Peptide Synthetases (NRPS) is a promising strategy for discovering new bioactive compounds, which can serve as valuable leads for drug development, such as new antibiotics. However, their engineering is hampered by the limited availability of molecular tools for the efficient heterologous expression of their large biosynthetic gene clusters. In fact, a single NRPS gene can already exceed the size limits of standard cloning vectors. In this study, we establish split inteins as a novel tool for NRPS engineering to enable the expression of single, covalently linked NRPS proteins from multiple plasmids and to perform cloning-free module swapping. Using the xenotetrapeptide synthetase as model system, we show that an NRPS can be split into three parts and reconstituted via trans-splicing using two orthogonal inteins at four different engineering sites. Based on this tripartite platform we build a library comprising 21 plasmids and generated 324 hybrid NRPS by combinatorial transformation. More than half were catalytically active, producing over 200 novel peptides. This intein-based technology provides a modular platform for generating natural product-like peptide libraries, expanding biocatalytically accessible chemical space.

|
Scooped by
?
October 3, 1:21 AM
|
Plant roots release exudates to encourage microbiome assembly, which influences the function and stress resilience of plants. How specific exudates drive spatial colonization patterns remains largely unknown. In this study, we demonstrate that endodermal Casparian strips—forming the root’s extracellular diffusion barrier—restrict nutrient leakage into the rhizosphere, coinciding with and controlling spatial colonization patterns of rhizobacteria. We find that vasculature-derived glutamine leakage is a major bacterial chemoattractant and enhancer of proliferation, defining a previously unknown pathway for root exudate formation. Bacteria defective in amino acid chemoperception display reduced attraction toward leakage sites, and roots with Casparian strip defects display bacterial overproliferation, dependent on bacterial capacity for amino acid metabolization. Associated chronic immune stimulation suggests that endodermal nutrient restriction is crucial for regulating microbial colonization and assembly, limiting excessive proliferation that could compromise plant health.
|

|
Scooped by
?
Today, 11:19 AM
|
Lysine acylations are a diverse class of post-translational modifications (PTMs) that dynamically regulate protein function. Access to homogenous, site-specifically modified proteins is essential for dissecting their molecular roles, yet remains challenging with traditional methods. Here we present a modular strategy that combines genetic code expansion (GCE) with a chemoselective amide bond-forming reaction to install a wide range of lysine acylations directly onto folded proteins. Key to this approach is the genetic encoding of Nϵ-methoxylysine, which reacts efficiently with N-methyliminodiacetyl (MIDA) acylboronates to generate diverse acyl lysine modifications from a single protein precursor, including several acyl PTMs not previously accessible through GCE. This mild and broadly applicable methodology enables systematic studies of lysine acylation on multiple target proteins. We demonstrate its utility by probing the effects of defined acyl modifications on enzymatic activity, protein-RNA interactions and deacylase-mediated regulation. Together, this platform establishes a versatile strategy to interrogate the functional consequences of this wide-spread class of PTMs.

|
Scooped by
?
Today, 10:40 AM
|
The structural integrity of bacterial cells is traditionally attributed to the peptidoglycan cell wall, and more recently to the outer membrane, with the cytoplasmic membrane assumed to be mechanically passive. Cells lacking filaments of the actin homolog MreB are more bendable, suggesting a role for the cytoskeleton in cell stiffness. Here, we show that MreB does not stiffen the envelope directly, but instead mechanically couples the cell wall to the cytoplasmic membrane through its role in peptidoglycan synthesis, increasing resistance to bending. Under hyperosmotic stress, MreB relocalized to the poles, forming linkages that prevent membrane detachment from the cell wall and attenuate cytoplasmic contraction. Disruption of MreB filament formation, nutrient starvation, or inactivation of glycan elongation factors abolished or reduced this coupling, revealing that peptidoglycan biosynthesis actively mediates stress distribution across surface layers. Our findings redefine the bacterial envelope as a mechanically integrated composite, with the cytoplasmic membrane having substantial load-bearing capacity.

|
Scooped by
?
October 3, 7:58 PM
|
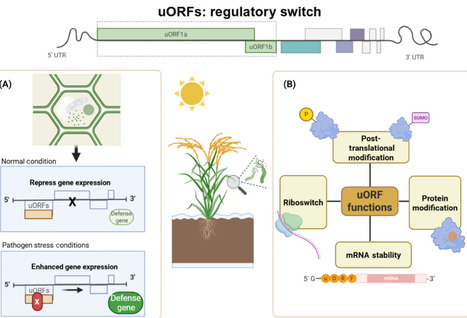
The discovery and manipulation of upstream open reading frames (uORFs) represent a promising frontier in plant biotechnology, offering strategies to enhance disease resistance and crop resilience. Here, we explore the role of uORFs in regulating gene expression under biotic stress and discuss approaches to engineer uORFs for sustainable agriculture and precision breeding.

|
Scooped by
?
October 3, 7:29 PM
|
The discovery of novel antimicrobial peptides (AMPs) against clinical superbugs is urgently needed to address the ongoing antibiotic resistance crisis. AMPs are promising candidates due to their broad-spectrum activity, rapid bactericidal mechanisms and reduced likelihood of inducing resistance compared with conventional antibiotics. Here, a pre-trained protein large language model (LLM), ProteoGPT, was established and further developed into multiple specialized subLLMs to assemble a sequential pipeline. This pipeline enables rapid screening across hundreds of millions of peptide sequences, ensuring potent antimicrobial activity and minimizing cytotoxic risks. Through transfer learning, we endowed the LLMs with different domain-specific knowledge to achieve high-throughput mining and generation of AMPs within a unified methodological framework. Notably, both mined and generated AMPs exhibited reduced susceptibility to resistance development in ICU-derived carbapenem-resistant Acinetobacter baumannii (CRAB) and methicillin-resistant Staphylococcus aureus (MRSA) in vitro. The AMPs also showed comparable or superior therapeutic efficacy in in vivo thigh infection mouse models compared with clinical antibiotics, without causing organ damage and disrupting gut microbiota. The mechanisms of action of these AMPs involve disruption of the cytoplasmic membrane and membrane depolarization. Overall, this study presents a generative artificial intelligence approach for the discovery of novel antimicrobials against multidrug-resistant bacteria, enabling efficient and extensive exploration of AMP space. This study presents a generative artificial intelligence approach for the high-throughput discovery of antimicrobials against multidrug-resistant bacteria.

|
Scooped by
?
October 3, 7:20 PM
|
Chromosome architecture plays a crucial role in bacterial adaptation, yet its direct impact remains unclear. Different bacterial species and even strains within the same species exhibit diverse chromosomal configurations, including a single circular or linear chromosome, two circular chromosomes, or a circular-linear combination. To investigate how these architectures shape bacterial behavior, we generated near-isogenic strains representing each configuration in Agrobacterium tumefaciens C58, an important soil bacterium widely used for plant genetic transformation. Strains with a single-chromosome architecture, whether linear or circular, exhibited faster growth, enhanced stress tolerance, and greater inter-strain competitiveness. In contrast, bipartite chromosome strains showed higher virulence gene expression and enhanced transient plant transformation efficiency, suggesting a pathogenic adaptation. Whole-transcriptome analysis revealed architecture-dependent gene expression patterns, underscoring the profound impact of chromosome organization on Agrobacterium fitness and virulence. These findings highlight how chromosome structure influences bacterial adaptation and shapes evolutionary trajectories.

|
Scooped by
?
October 3, 7:00 PM
|
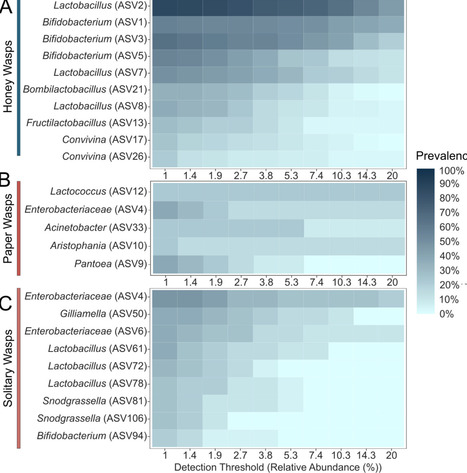
Honey-feeding social bees, including honey bees, bumble bees, and stingless bees, possess distinctive gut bacterial communities that provide benefits to hosts, such as defense against pathogens and parasites. Members of these communities are transmitted through social interactions within colonies. The Mexican honey wasp (Brachygastra mellifica) represents an independent origin of honey-storing within a group of social Hymenoptera. Honey wasps feed on and store honey, but, unlike bees, they prey on other insects as a protein source and do not consume pollen. We surveyed the gut bacterial communities of Mexican honey wasps across sites within Texas using 16S ribosomal RNA profiling, and we estimated bacterial titer per bee using quantitative PCR. For comparison, we also surveyed non-honey-feeding wasps from six families, collected in the same region. We found that honey wasp communities are dominated by characteristic bacterial species. In contrast, other wasps had lower absolute titers and more variable communities, dominated by environmental bacteria. Honey wasps from all sampled nests contained strains of Bifidobacterium and Bombilactobacillus that were closely related to symbionts of bumble bees and other bees, suggesting acquisition via host-switching. Some individuals also harbored a close relative of Candidatus Schmidhempelia bombi (Orbaceae), an uncultured bumble bee symbiont, again suggesting host-switching. The most prevalent species was an uncultured Lactobacillus that potentially represents an independent acquisition of environmental Lactobacillus. The transition to honey feeding, combined with a highly social life history, appears to have facilitated the establishment of a bacterial community with similarities to those of social bees.

|
Scooped by
?
October 3, 5:11 PM
|
Uncovering genotype-phenotype relationships is hampered by genetic redundancy. For example, most genes in Streptococcus pneumoniae are non-essential under laboratory conditions. A powerful approach to unravel genetic redundancy is by identifying gene-gene interactions. We developed a broadly applicable dual CRISPRi-seq method and analysis pipeline to probe genetic interactions (GIs) genome-wide. A library of 869 dual single-guide RNAs (sgRNAs) targeting high-confidence operons was created, covering over 70% of the genetic elements in the pneumococcal genome. Testing these 378,015 unique combinations, 4,026 significant GIs were identified. Besides known GIs, we found previously unknown positive and negative interactions involving genes in fundamental cellular processes such as division and chromosome segregation. The presented methods and bioinformatic approaches can serve as a roadmap for genome-wide gene interaction studies in other organisms. All interactions are available for exploration via the Pneumococcal Genetic Interaction Network (PneumoGIN), which can serve as a starting point for new biological discoveries.

|
Scooped by
?
October 3, 12:32 PM
|
Reactive oxygen species (ROS), inevitable byproducts of aerobic respiration, exhibit both signaling and cytotoxic effects depending on concentration within cells. At controlled levels, ROS serve as crucial signaling molecules mediating intracellular signal transduction pathways. However, excessive accumulation triggers oxidative stress, leading to oxidative damage that disrupts protein structure and function, ultimately inducing cell death. Enhancing cellular adaptive responses to oxidative stress and mitigating intracellular ROS accumulation are therefore critical strategies for maintaining cellular homeostasis. This approach significantly improves product yield and underpins its widespread application across diverse sectors, including functional foods, pharmaceuticals, and cosmeceuticals. This review first delineates the origins and multifaceted roles of ROS. Subsequently, it details strategies employed to bolster cellular oxidative stress resilience, focusing on process optimization and metabolic engineering approaches. Finally, synthesizing current research advances, existing challenges, and emerging trends, the review outlines future research directions aimed at enhancing the oxidative stress tolerance of microbial cell factories.

|
Scooped by
?
October 3, 10:43 AM
|
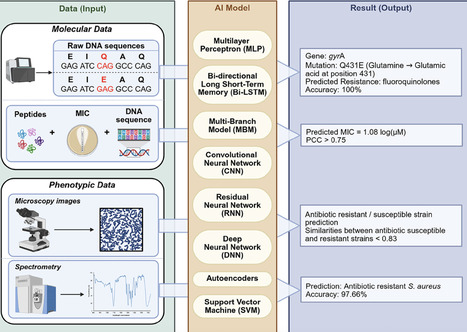
Antibiotic resistance continues to erode the effectiveness of modern medicine, creating an urgent demand for rapid and reliable diagnostic solutions. Conventional diagnostic approaches, including culture-based susceptibility testing, remain the clinical reference standard but are constrained by lengthy turnaround times and limited sensitivity for early detection. In recent years, significant progress has been made with molecular and spectrometry-based methods, such as PCR and next-generation sequencing, MALDI-TOF MS, Raman and FTIR spectroscopy, alongside emerging CRISPR-based platforms. Complementary innovations in biosensors, microfluidics, and artificial intelligence further expand the diagnostic landscape, enabling faster, more sensitive, and increasingly portable assays. This review examines both established and emerging technologies for detecting antibiotic resistance, outlining their respective strengths, limitations, and potential roles across diverse settings. By synthesizing current advances and highlighting future opportunities, this review emphasizes complementarities among detection strategies and their potential integration into practical diagnostic frameworks, including in resource-limited settings.

|
Scooped by
?
October 3, 10:31 AM
|
Growing environmental concerns and the short lifespan of synthetic polymers have intensified the search for sustainable, biodegradable alternatives. Microbially produced biopolymers, such as bacterial cellulose, hyaluronic acid, polyhydroxyalkanoates, and poly-γ-glutamic acid, have gained attention due to their biocompatibility, non-toxicity, and renewability. This review highlights recent advances in metabolic engineering aimed at improving the yield, quality, and functionality of these biopolymers. Key strategies include genetic modifications (e.g., gene overexpression, gene deletion, CRISPR-Cas9) and systems biology tools (e.g., proteomics, genomics, synthetic biology). These approaches enable the development of optimized materials for food, medical, and industrial applications. The review also addresses important factors such as molecular weight control, scalability, biocompatibility, and eco-friendly degradation. By comparing natural and engineered microbial platforms, we provide insights into their advantages and limitations. This work underscores the crucial role of microbial metabolic engineering in advancing next-generation biopolymers, supporting both industrial innovation and global sustainability goals.

|
Scooped by
?
October 3, 10:24 AM
|
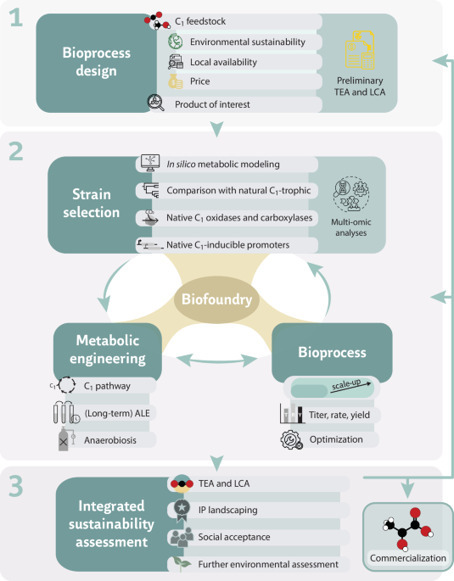
Engineering microbes for synthetic one-carbon (C1) assimilation continues to gain momentum with the expanding demand for sustainable bioprocesses. While most efforts focused on model microbes, non-canonical hosts offer untapped potential due to native metabolic properties, enzyme activities, and substrate tolerance. This perspective outlines key considerations for selecting and engineering such strains, including metabolic modeling, use of native C1-inducible promoters, and adaptation to anaerobic conditions. Environmental impacts are evaluated through life cycle assessment, identifying substrates with low carbon footprints. Integrating techno-economic and sustainability insights at early stages is essential to guide the development of efficient, scalable C1-based biomanufacturing systems. Synthetic one-carbon assimilation could contribute to a more sustainable and circular carbon economy, but much work in this field has focused on model microorganisms. Here the authors provide their perspective on the potential value of non-model microbes, and how that potential could be realised.

|
Scooped by
?
October 3, 1:21 AM
|
Advances in artificial intelligence (AI)–assisted protein engineering are enabling breakthroughs in the life sciences but also introduce new biosecurity challenges. Synthesis of nucleic acids is a choke point in AI-assisted protein engineering pipelines. Thus, an important focus for efforts to enhance biosecurity given AI-enabled capabilities is bolstering methods used by nucleic acid synthesis providers to screen orders. We evaluated the ability of open-source AI-powered protein design software to create variants of proteins of concern that could evade detection by the biosecurity screening tools used by nucleic acid synthesis providers, identifying a vulnerability where AI-redesigned sequences could not be detected reliably by current tools. In response, we developed and deployed patches, greatly improving detection rates of synthetic homologs more likely to retain wild type–like function.
|
 Your new post is loading...
Your new post is loading...