Your new post is loading...

|
Scooped by
?
Today, 12:52 AM
|
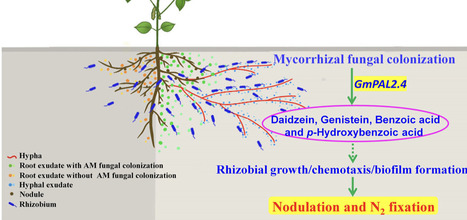
Legume plants commonly associate with both arbuscular mycorrhizal (AM) fungi and rhizobia and thus enhance the acquisition of phosphorus (P) and nitrogen (N) nutrition. Inoculation with AMF can promote nodulation and N2 fixation of legume plants; however, the underlying mechanisms remain poorly understood. Here, root exudates collected from AM-colonised soybean plants showed greater accumulation of the specific flavonoids (daidzein and genistein) and phenolic acids (benzoic acid and p-Hydroxybenzoic acid), and significantly promoted nodulation. Furthermore, the exudates from AM-colonized roots and the derived specific flavonoids and phenolic acids effectively increased rhizobial growth, chemotaxis, biofilm formation. Addition of the specific synthetic root exudates enhanced nodulation and N2 fixation, and expression of the core nodulation genes in soybean. Overexpression of a phenylalanine ammonia-lyase gene, GmPAL2.4 markedly upregulated the expression of the genes related to the biosynthesis of daidzein, genistein, benzoic acid, and p-Hydroxybenzoic acid, and increased accumulation of these specific flavonoids and phenolic acids in the transgenic plants, thus enhancing nodulation and N2 fixation. In summary, we demonstrated a crucial role of specific flavonoids and phenolic acids induced by AM symbiosis in promoting rhizobium-host symbiosis. This offers a pathway for improving symbiotic efficiency through the use of specific synthetic compounds.

|
Scooped by
?
Today, 12:39 AM
|
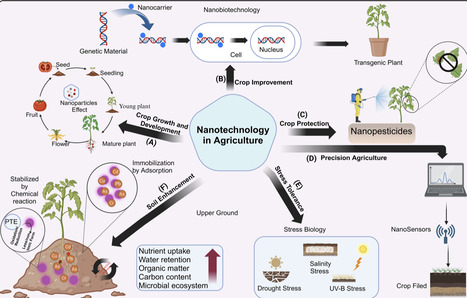
The rising global demand for food poses a significant threat to environmental health through both biotic (e.g., pests, pathogens) and abiotic (e.g., drought, salinity) stresses. Therefore, the adoption of innovative strategies is essential to ensure the sustainability of agricultural practices and to enhance crop resilience against environmental challenges. This review investigates how the integration of nanotechnology with genetically modified (GM) crops can offer solutions to agricultural challenges by improving crop resilience and productivity. While genetic modification has faced limitations in achieving consistent results due to environmental variability and species-specific differences, nanotechnology has emerged as a transformative tool to enhance GM crop performance. In this study we critically explore the underlying mechanisms of combining nanotechnology with GM crops to enhance plant growth and development and their resilience against biotic and abiotic stresses. Furthermore, nanotechnology also play a crucial role in targeted gene delivery, precise genome editing, and controlled regulation of gene expression in GM plant cells. Overall, the emerging role of nanotechnology in GM crops is paving the way for innovative solutions in agriculture. By leveraging nanotechnology, researchers are exploring novel approaches to enhance productivity, combat plant diseases, and improve plant resilience to environmental stress for sustainable agriculture. Furthermore, in this review we also highlighted the environmental impacts and safety issues associated with using nanotechnology in crops in order to establish more resilient and sustainable farming practices.

|
Scooped by
?
Today, 12:21 AM
|
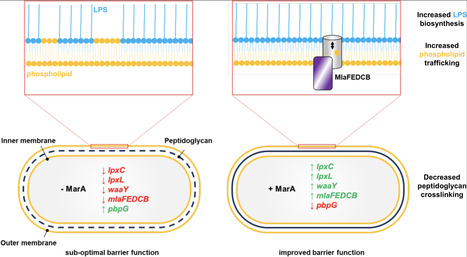
The multiple antibiotic resistance activator (MarA) protein is a transcription factor implicated in control of intrinsic antibiotic resistance in enteric bacterial pathogens. In this work, we screened the Escherichia coli genome computationally for MarA binding sites. By incorporating global maps of transcription initiation, and clustering predicted targets according to gene function, we were able to avoid widespread misidentification of MarA sites, which has hindered prior studies. Subsequent genetic and biochemical analyses identified direct activation of genes for lipopolysaccharide (LPS) biosynthesis and repression of a cell wall remodelling endopeptidase. Rewiring of the MarA regulon, by mutating subsets of MarA binding sites, reveals synergistic interactions between regulatory targets of MarA. Specifically, we show that uncoupling LPS production, or cell wall remodelling, from regulation by MarA, renders cells hypersensitive to mutations altering lipid trafficking by the MlaFEDCB system. Together, our findings demonstrate how MarA co-regulates different aspects of cell envelope biology to maximise antibiotic resistance.

|
Scooped by
?
May 5, 11:59 PM
|
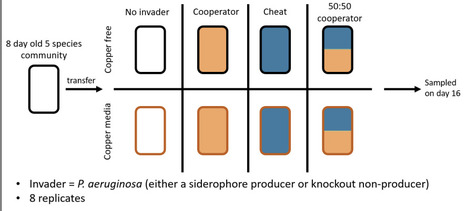
Successful biological invasions are dependent on the invader being able to grow and reproduce in the new environment. One way that microbial invaders may facilitate this is to use cooperative public goods, such as metal-binding siderophores. However, siderophore production can be exploited by non-producing cheats who benefit from production without paying any associated costs. Here, we test the importance of cooperation for the success of Pseudomonas aeruginosa invading a 5-species microbial community. We do this by comparing the success of a siderophore-producing strain, a siderophore-deficient mutant strain and a 50:50 mixed population, both in environments with weak (copper absent) and strong (copper present) siderophore requirement. We found no effect of invader type on success when siderophores were less essential for growth, but large differences when they were selectively favoured. Here the producer-cheat mix had the greatest success, with both strains having near equal fitness and reaching high densities, whilst in isolation producers had intermediate success and cheats the lowest. Similarly, resident diversity only differed across invader treatments when copper was present. In conclusion, we show that the presence of cheats can provide a larger benefit for invasion success than pure cooperator populations, but only when public goods are particularly beneficial.

|
Scooped by
?
May 5, 11:03 PM
|
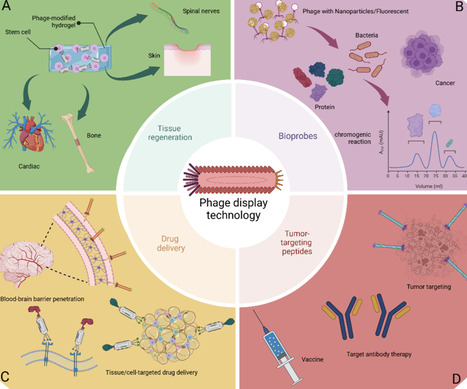
Phage display technology, a molecular screening tool introduced in 1985, facilitates the display of peptides or proteins on phage surfaces to identify high-affinity ligands. Phage display has enabled the identification of peptides that enhance biomaterial biocompatibility, support stem cell proliferation and differentiation, and facilitate targeted drug delivery. Additionally, it has been utilized in bacterial detection, disease biomarker identification, and targeted therapies, including crossing the blood–brain barrier for improved cancer treatments. Despite challenges such as peptide stability and immunogenicity, ongoing innovations continue to enhance its therapeutic potential. This review discusses the potential of phage display technology for empowering healthcare through demonstrating its development in various biological fields in recent years.

|
Scooped by
?
May 5, 10:58 PM
|
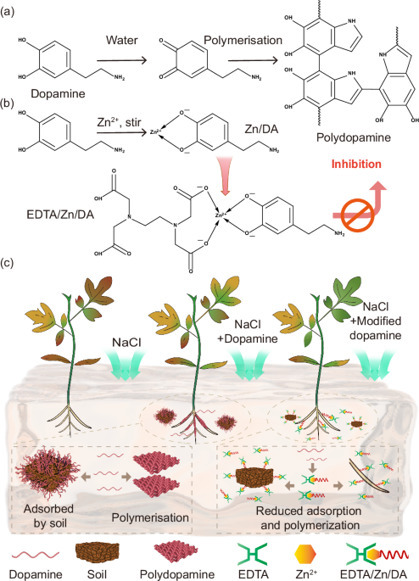
Soil salinization hinders sustainable development of global agriculture. Dopamine (DA) delivery is promising for mitigating the detrimental effects of salt on plants. However, self-polymerization limits delivery and effectiveness. Here we chelated DA with ethylenediamine tetraacetic acid and zinc to reduce self-polymerization. To reduce soil adsorption, a sodium lignosulfonate and octadecyl dimethyl benzyl ammonium chloride nanocarrier is made for delivery to the plant. Compared with DA monomer, the soil adsorption rate of the DA in the nanocarrier is 46.02% lower. Salt stress experiments reveal, compared with NaCl and DA groups, the nanocarrier group exhibits significant increases in growth indicators for tomato plants. The beneficial effect is attributed to the increases in proline content, antioxidant capacity, and K+/Na+ ratios in the plants. Similar results are also observed with woody pear seedlings. These findings provide insights into alleviating crop salt stress. Soil salinisation hinders sustainable development of global agriculture. Here the authors report on a carrier designed to delivery dopamine to plants to improve salt resistance, avoiding dopamine self-polymerisation and soil adsorption, demonstrating increased tomato and pear seedling growth under salt stress.

|
Scooped by
?
May 5, 10:25 PM
|
Molecular cloning through in vivo assembly (IVA) is an efficient homology-based approach that can achieve complex cloning operations in a single step, bypassing the need for enzymes and reducing hands-on time and costs. However, primer design remains the most demanding task, requiring substantial expertise and a source of human error. Here, we introduce “IVA Prime,” an intuitive online tool for automated primer design for IVA cloning, which reduces the workload and avoids errors. We describe how IVA Prime generates optimized primers for all cloning operations, i.e. mutagenesis, insertions, deletions, and subcloning. IVA Prime provides an intuitive interface and robust algorithms, thus mitigating the complexities associated with primer design into a single mouse click. A set of thoroughly tested default parameters allows both inexperienced users and experts to generate ready-to-order primer sequences for a standardized cloning protocol with high success rate. We describe the method and demonstrate its efficiency and reliability with example applications. IVA Prime is freely available at https://www.ivaprime.com.

|
Scooped by
?
May 5, 9:47 PM
|
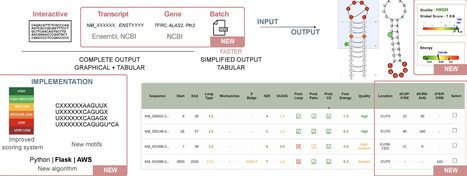
Iron-responsive elements (IREs) are cis-acting regulatory RNA motifs that bind to iron regulatory proteins (IRP1 and IRP2), playing an essential role in the post-transcriptional regulation of genes involved in iron metabolism. Disruptions in this IRP/IRE regulatory system have been linked to several human diseases. SIREs (searching for IREs) is a web-server tool designed to predict IREs in nucleotide sequences. Here, we present SIREs 3.0 webserver, an improved new version built on a Flask-based framework, replacing the previous Perl backend to improve interconnectivity with other services. This upgrade introduces three novel input methods—batch, transcript, and gene modes—to cover researchers’ needs, from large-scale analysis to single-gene queries. By integrating with NCBI and Ensembl APIs, SIREs 3.0 fetches genomic data to improve predictions with novel features, such as the location of the IREs in the transcript. Other novelties include novel IRE motifs based on in vivo verified data, broadening the scope of IRE detection. The scoring system has been refined with empirical data and enhanced graphical representations of predicted IREs have been added to the output. The interface is now more responsive and accessible across all devices. SIREs 3.0 can be accessed at: https://www.sires-webserver.eu.

|
Scooped by
?
May 5, 9:05 PM
|
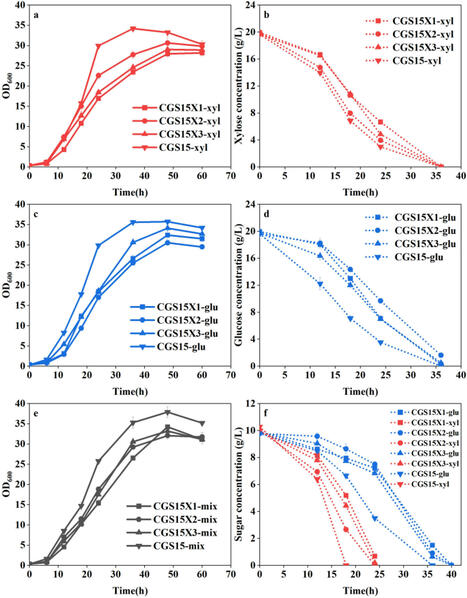
To fully utilize lignocellulosic hydrolysate abundant in xylose content, it is necessary to engineer a Corynebacterium glutamicum strain that can preferentially and efficiently utilize xylose in the presence of glucose/xylose mixtures. C. glutamicum strain CGS15X5-E2 was obtained through metabolic engineering and adaptive laboratory evolution (ALE), which preferentially utilizes xylose and completely consumes it before switching to glucose utilization. A genetically defined chassis strain, CGS15X57, was constructed to switch to glucose consumption only after xylose was depleted based on genome analysis and mutation reconstruction, in which xylose utilization capability was also enhanced. The average xylose consumption rate of CGS15X57 reached 0.833 ± 0.048 g/l/h, which was 28.0% higher than that of the control. Three new beneficial mutations (Cgl1992267 insert, Cgl2948G208 T and Cgl2948556△C) endow C. glutamicum with rapid growth and efficient xylose utilization phenotypes. A chassis strain of C. glutamicum that preferentially and efficiently utilizes xylose has been obtained, facilitating the full utilization of lignocellulosic hydrolysates and the construction of co-culture systems under glucose/xylose mixed sugar conditions.

|
Scooped by
?
May 5, 12:24 AM
|
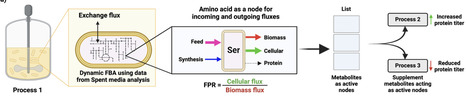
Complex media supplemented with a carbon source are commonly used in bioprocesses for recombinant protein production in Escherichia coli. Optimizing these processes is challenging and requires precise understanding of cellular metabolism and nutrient requirements. Compared to a design of experiments approach that necessitates extensive experimentation, metabolic modeling using a genome scale metabolic model (GEM) offers a more predictive and systematic approach to guide process optimization by identifying specific metabolic bottlenecks. In addition, spent media analysis (SMA) can unravel the preferential utilization of different media components during the bioprocess. Here, we integrated the updated E. coli GEM with time course SMA data from a fed-batch process and performed dynamic flux balance analysis (dFBA) to identify metabolites that function as active nodes and are vital for cellular function. These are potential target supplements to boost cellular activity and in turn the recombinant protein productivity. Using an iterative approach of performing fermentation, SMA, and metabolic modeling, we intensified the bioprocess in just five experimental trials, resulting in a six-fold increase in protein productivity. Our new feeding strategy involved yeast extract with amino acid supplementation (Ser, Thr, Asp, and Glu) and increased oxygen transfer rates. This approach demonstrates significant promise for application in bioprocess intensification.

|
Scooped by
?
May 5, 12:09 AM
|
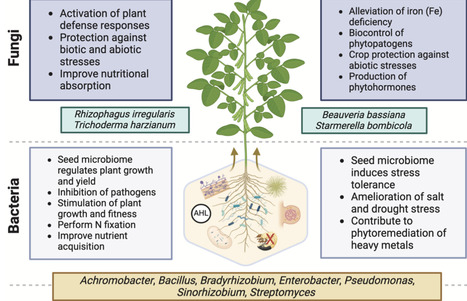
In the context of global challenges such as climate change, soil degradation, and food security, understanding the modes of action of Plant Growth-Promoting Microorganisms (PGPMs), their formulation, and their application is crucial and can be more focused in future research projects. This editorial paper aims to elucidate diverse modes of action employed by different types of PGPMs, including nitrogen fixation, phosphorus solubilization, production or regulation of phytohormones, and plant protection against environmental and biotic stresses as demonstrated and discussed in the Special Issue entitled “Plant Growth-Promoting Microorganisms: new insights and the way forward”.

|
Scooped by
?
May 4, 10:58 PM
|
Aspergillus fumigatus is a saprophytic fungus prevalent in the environment and capable of causing severe invasive infection in humans. This organism can use strategies such as molecule masking, immune response manipulation and gene expression alteration to evade host defences. Understanding these mechanisms is essential for developing effective diagnostics and therapies to improve patient outcomes in Aspergillus-related diseases. In this Review, we explore the biology and pathogenesis of A. fumigatus in the context of host biology and disease, highlighting virus-associated pulmonary aspergillosis, a newly identified condition that arises in patients with severe pulmonary viral infections. In the post-pandemic landscape, in which immunotherapy is gaining attention for managing severe infections, we examine the host immune responses that are critical for controlling invasive aspergillosis and how A. fumigatus circumvents these defences. Additionally, we address the emerging issue of azole resistance in A. fumigatus, emphasizing the urgent need for greater understanding in an era marked by increasing antimicrobial resistance. This Review provides timely insights necessary for developing new immunotherapeutic strategies against invasive aspergillosis. In this Review, van de Veerdonk and colleagues explore Aspergillus fumigatus biology, the host immune responses and the fungal mechanisms of immune evasion, as well as emerging challenges such as virus-associated pulmonary aspergillosis and azole resistance, and reflect on future directions for therapeutics and patient care improvement.

|
Scooped by
?
May 4, 10:47 PM
|
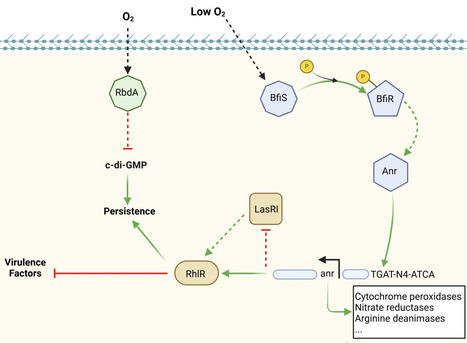
The exquisite ability of bacteria to adapt to their environment is essential for their capacity to colonize hostile niches. In the cystic fibrosis (CF) lung, hypoxia is among several environmental stresses that opportunistic pathogens must overcome to persist and chronically colonise. Although the role of hypoxia in the host has been widely reviewed, the impact of hypoxia on bacterial pathogens has not yet been studied extensively. This review considers the bacterial oxygen-sensing mechanisms in three species that effectively colonise the lungs of people with CF, namely Pseudomonas aeruginosa, Burkholderia cepacia complex and Mycobacterium abscessus and draws parallels between their three proposed oxygen-sensing two-component systems: BfiSR, FixLJ, and DosRS, respectively. Moreover, each species expresses regulons that respond to hypoxia: Anr, Lxa, and DosR, and encode multiple proteins that share similar homologies and function. Many adaptations that these pathogens undergo during chronic infection, including antibiotic resistance, protease expression, or changes in motility, have parallels in the responses of the respective species to hypoxia. It is likely that exposure to hypoxia in their environmental habitats predispose these pathogens to colonization of hypoxic niches, arming them with mechanisms than enable their evasion of the immune system and establish chronic infections. Overcoming hypoxia presents a new target for therapeutic options against chronic lung infections.
|

|
Scooped by
?
Today, 12:47 AM
|
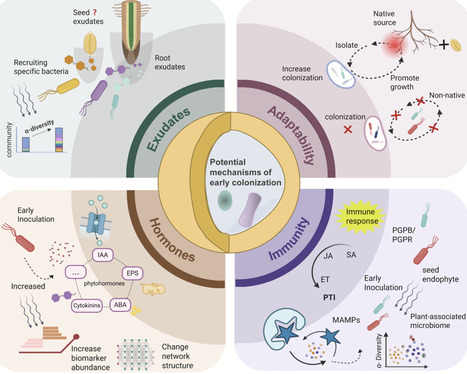
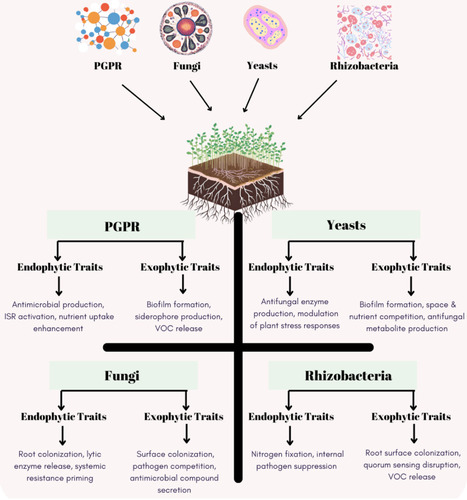
The relationship between plants and early colonizing microbes is crucial for regulating agricultural ecosystems. Recent evidence strongly suggests that by introducing beneficial microbes during the seed or seedling stages, the diversity and assembly structure of the plant-related microbial community during later plant development can be altered, recruiting beneficial bacteria to enhance plant protection. However, the mechanisms of community assembly and their effects on plant growth are still not fully understood. To deepen our understanding of the importance of early inoculation for improving plant performance, this review comprehensively summarizes recent research advancements on the effects of early introduction on plant growth and adaptability. The mechanisms and ecological significance of early inoculation in the assembly of plant-related bacterial communities are discussed, with particular emphasis on the importance of seed endophytes, plant growth-promoting rhizobacteria (PGPR), and synthetic microbial consortia as microbial inoculants in enhancing plant health and productivity. Additionally, this review proposes a new strategy: sequential inoculation during the seed and seedling stages, aiming to maximize the effects of microbes.

|
Scooped by
?
Today, 12:35 AM
|
Ribosomal DNA (rDNA) in eukaryotes is maintained in hundreds of copies with rDNA copy number varying greatly among individuals within a species. In the budding yeast Saccharomyces cerevisiae, the rDNA copy number across wild isolates ranges from 90 to 300 copies. Previous studies showed that 35 rDNA copies are sufficient for ribosome biogenesis in this yeast and enable wild-type-like growth in standard laboratory growth conditions. We addressed two major questions concerning rDNA copy number variation in this yeast: (1) What are the fitness consequences of rDNA copy number variation outside and within the natural range in standard laboratory growth conditions? (2) Do these fitness effects change in different growth conditions? We used growth competitions to compare the fitness effects of rDNA copy number variation in otherwise isogenic strains whose rDNA copy number ranged from 35 to 200. In standard growth conditions, we found that fitness gradually increases from 35 rDNA copies until reaching a plateau that spans from 98 to 160 rDNA copies, well within the natural range. However, rDNA copy number-dependent fitness differed across environments. The gradual fitness increase with increasing rDNA copy number in standard growth conditions gave way to a markedly lower fitness of strains with copy numbers below the natural range in these two stress conditions. These results suggest that selective pressures drive rDNA copy number in this yeast to at least ∼100 copies and that a higher number of copies might buffer against environmental stress. The similarity of the S. cerevisiae rDNA copy number range to the ranges reported in C. elegans, D. melanogaster, and humans points to conserved selective pressures maintaining the range of natural rDNA copy number in these highly diverse species.

|
Scooped by
?
Today, 12:04 AM
|
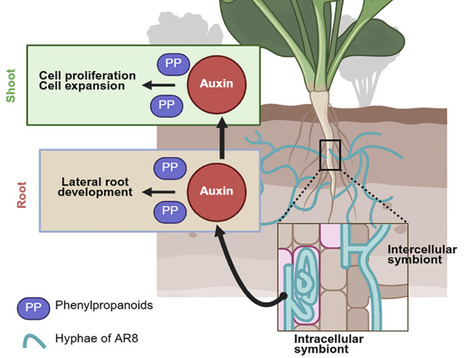
Beneficial association with symbiotic fungi helps improve growth and fitness in most land plants and shows great potential as biofertilizers in precision agriculture. Here, we demonstrated that a root fungal endophyte, Tinctoporellus species isolate AR8, enabled yield improvement in Brassicaceae leafy green choy sum (Brassica rapa var. parachinensis). Mechanistically, AR8 colonized the root cortex/endosphere and channeled the metabolic flux to phenylpropanoids and requisite secondary metabolites to promote plant growth. AR8-assisted biosynthesis of auxin improved root growth and provided an intrinsic source for long-distance signaling that enhanced shoot biomass. Chemical complementation with exogenous p-coumaric acid restored auxin signaling and enhanced growth in AR8-inoculated pal1 mutant plants, thus implicating such a phenylpropanoid-auxin nexus as a pivotal regulator of symbiotic plant growth. Comparative metabolomics established hydroxycinnamic acid and p-coumaric acid as major plant-growth-promoting hubs that bridge the phenylpropanoid pathway and auxin signaling in the cross-kingdom AR8 symbiotic interaction model.

|
Scooped by
?
May 5, 11:36 PM
|
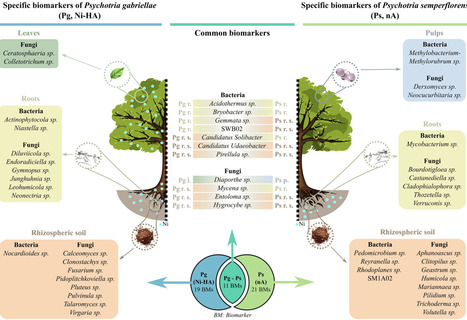
In New Caledonia, nearly 2000 plant species grow on ultramafic substrates, which contain prominent levels of heavy metals and are deficient in essential plant nutrients. To colonize these habitats, such plants, known as metallophytes, have developed various adaptive behaviors towards metals (exclusion, tolerance, or hyperaccumulation). Ultramafic substrates also host many unique microorganisms, which are adapted to metallic environments and capable of boosting plant growth while assisting plants in acquiring essential micronutrients. Hence, plant-microbiota interactions play a key role in adapting to environmental stress. Here, we hypothesized that microbial associations in the different aboveground and underground compartments of metallophytes could be associated to their metal hyperaccumulation or exclusion phenotypes. This hypothesis was tested using a systematic comparative metabarcoding approach on the different compartments of two New Caledonian metallophytes belonging to the same genus and living in sympatry on ultramafic substrates: Psychotria gabriellae, a nickel-hyperaccumulator (Ni-HA), and Psychotria semperflorens, the related non-accumulator (nA) species. The study of the diversity and specificity of fungal amplicon sequence variants (ASVs) reveals a structuring of fungal communities at both the plant phenotype and compartment levels. In contrast, the structure of bacterial communities was primarily shaped by the belowground compartments. Additionally, we observed a lower diversity in the bacterial communities of the aboveground compartments of each species. For each plant species, we highlighted a distinct global microbial signature (biomarkers), as well as compartment-specific microbial associations. To our knowledge, this study is the first to systematically compare the microbiomes associated with different compartments of New Caledonian metallophyte species growing on the same substrate and under identical environmental conditions but exhibiting different adaptive phenotypes. Our results reveal distinct microbial biomarkers between the Ni-hyperaccumulator and non-accumulator Psychotria species. Most of the highlighted biomarkers are abundant in various plants under metal stress and may contribute to improving the phytoextraction or phytostabilization processes. They are also known to tolerate heavy metals and enhance metal stress tolerance in plants. The present findings highlight that the microbial perspective is essential for better understanding the mechanisms of hyperaccumulation and exclusion at the whole-plant level.

|
Scooped by
?
May 5, 11:00 PM
|
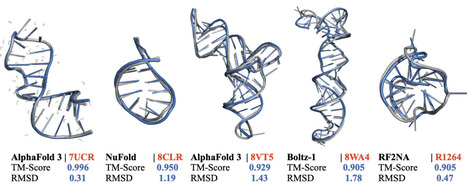
In recent years, tremendous advances have been made in predicting protein structures and protein-protein interactions. However, progress in predicting the structure of RNA, either alone or in complex with other macromolecules, has been less prominent, although some recent developments have been reported. It remains unclear whether the improved prediction accuracy is sustained for novel RNA structures. Results: Here, we use an independent benchmark to evaluate the performance of the latest methods. First, we show that state-of-the-art methods can sometimes predict the structure of single-chain RNA strands, with accurate models observed for RNAs with well-defined or regular secondary structures. Next, our evaluation was extended to RNA complexes, where prediction accuracy was notably higher for those involving extensive canonical base pairing. Additionally, a structural similarity analysis revealed that prediction success strongly correlates with resemblance to known structures, indicating that current methods recognise recurring motifs rather than generalising to novel folds. Finally, we also noted that the estimation of accuracies for RNA models is far from accurate. Therefore, it is not possible to reliably identify the correctly predicted models with today's methods. Availability and Implementation: Code used for running the methods and analyzing the main results is available from https://github.com/iammarcol/RNA-Benchmark and here https://zenodo.org/records/15304777

|
Scooped by
?
May 5, 10:33 PM
|
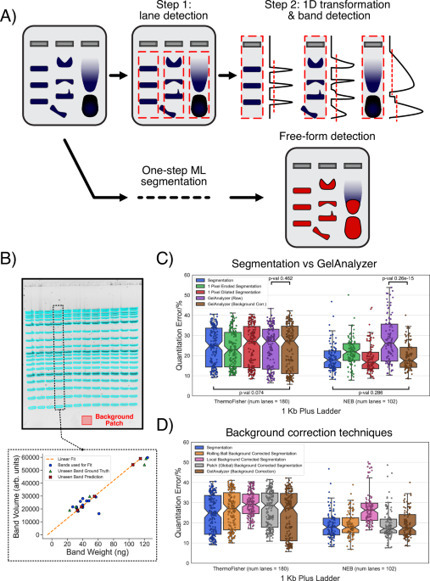
Gel electrophoresis is a ubiquitous laboratory method for the separation and semi-quantitative analysis of biomolecules. However, gel image analysis principles have barely advanced for decades, in stark contrast to other fields where AI has revolutionized data processing. Here, we show that an AI-based system can automatically identify gel bands in seconds for a wide range of experimental conditions, surpassing the capabilities of current software in both ease-of-use and versatility. We use a dataset containing 500+ images of manually-labelled gels to train various U-Nets to accurately identify bands through segmentation, i.e. classifying pixels as ‘band’ or ‘background’. When applied to gel electrophoresis data from other laboratories, our system generates results that quantitatively match those of the original authors. We have publicly released our models through GelGenie, an open-source application that allows users to extract bands from gel images on their own devices, with no expert knowledge or experience required. Gel electrophoresis image analysis still largely relies on manual or semi-automatic tools, limiting both efficiency and reproducibility. Here, authors introduce GelGenie, an AI-driven open-source platform that rapidly detects gel bands under various conditions.

|
Scooped by
?
May 5, 9:50 PM
|
The data resources provided by the European Bioinformatics Institute (EMBL-EBI) cover major areas of biological and biomedical research, giving free and open access to users ranging from expert to casual level. The EBI Search engine provides a unified metadata search engine across these resources. It provides a full-text search engine across over 6.5 billion data items, accessed through a user-friendly website and an OpenAPI-compliant programmatic interface. Here, we discuss recent developments and improvements in the service.

|
Scooped by
?
May 5, 9:43 PM
|
The escalating threats posed by plant pathogens and the environmental repercussions of conventional agrochemicals necessitate sustainable agricultural solutions. This review focuses on plant growth-promoting microorganisms (PGPR) such as bacteria, filamentous fungi, and yeasts, which play a pivotal role as biocontrol agents. These organisms enhance plant growth and resilience through nutrient solubilization, phytohormone production, and antagonistic activities against pathogens, offering a dual benefit of disease suppression and growth enhancement. However, the effective application of PGPMs faces challenges, including variability in field performance, survival and colonization under field conditions, and regulatory hurdles. This paper discusses these challenges and explores recent advances in utilizing these bioagents in sustainable agriculture, underscoring the importance of integrated pest management systems that reduce chemical inputs, thus promoting ecological balance and sustainable farming practices.

|
Scooped by
?
May 5, 12:31 AM
|
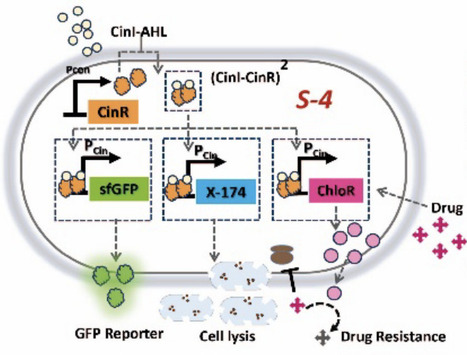
Synthetic biology enables the precise control of bacterial population dynamics through engineered genetic circuits, paving the way for innovative therapeutic and biosensing applications. Here, we present genetic circuit design that leverages AHL (acyl-homoserine lactone)-based quorum sensing and a transcriptional cascade regulated by an external inducer to drive synchronized proliferation and lysis events. In this system, the absence of the inducer arabinose prevents CinR mediated repression, enabling activation of a transcriptional cascade that drives the expression of the lysis protein, fluorescent protein, and a drug resistance gene. The accumulation of AHL acts as a proxy for population density and modulates gene expression through a quorum-sensing mechanism. This inducible circuit provides tunable and pulsatile control over bacterial population dynamics, enabling cycles of population growth and collapse. Such behavior prevents premature population clearance while ensuring functional bacterial densities for extended durations. Our system represents a foundational step toward implementing more complex multi-stable architectures for self-regulation of bacterial population. This dynamic and modular framework offers significant advantages for therapeutic interventions, such as self-regulating drug delivery, inflammation-responsive probiotic therapies, and targeted bacterial therapy.

|
Scooped by
?
May 5, 12:13 AM
|
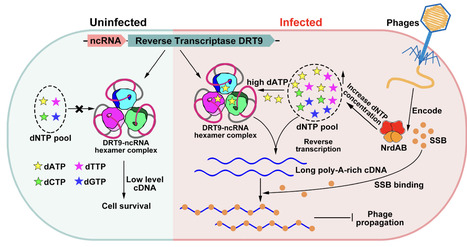
Prokaryotic defense-associated reverse transcriptases (DRTs) were recently identified with antiviral functions; however, their functional mechanisms remain largely unexplored. Here we show that DRT9 forms a hexameric complex with its upstream non-coding RNA (ncRNA) to mediate antiphage defense by inducing cell growth arrest via abortive infection. Upon phage infection, the phage-encoded ribonucleotide reductase NrdAB complex elevates intracellular dATP levels, activating DRT9 to synthesize long, poly-A-rich single-stranded cDNA, which likely sequesters the essential phage SSB protein and disrupts phage propagation. We further determined the cryo-electron microscopy structure of the DRT9-ncRNA hexamer complex, providing mechanistic insights into its cDNA synthesis. These findings highlight the diversity of RT-based antiviral defense mechanisms, expand our understanding of RT biological functions, and provide a structural basis for developing DRT9-based biotechnological tools.

|
Scooped by
?
May 4, 11:02 PM
|
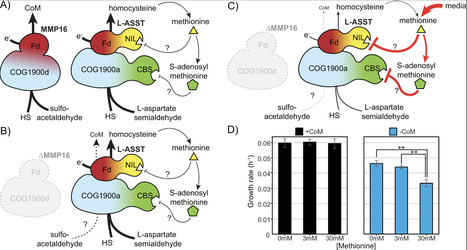
2-mercaptoethanesulfonate (Coenzyme M, CoM) is an organic sulfur-containing cofactor used for hydrocarbon metabolism in archaea and bacteria. In archaea, CoM serves as an alkyl group carrier for enzymes belonging to the alkyl-CoM reductase family, including methyl-CoM reductase, which catalyzes methane formation in methanogens. Two pathways for the biosynthesis of CoM have been identified in methanogenic archaea. The initial steps of these pathways are distinct but the last two reactions, leading up to CoM formation, are universally conserved. The final step is proposed to be mediated by methanogenesis marker metalloprotein 16 (MMP16), a putative sulfurtransferase, that replaces the aldehyde group of sulfoacetaldehyde with a thiol to generate CoM. Based on prior research, assignment of MMP16 as CoM synthase (ComF) is not widely accepted as deletion mutants have been shown to grow without any CoM dependence. Here, we investigate the role of MMP16 in the model methanogen, Methanosarcina acetivorans. We show that a mutant lacking MMP16 has a CoM-dependent growth phenotype and a global transcriptomic profile reflective of CoM-starvation. Additionally, the ∆ MMP16 mutant is a CoM auxotroph in sulfide-free medium. These data reinforce prior claims that MMP16 is a bona fide ComF but point to backup pathway(s) that can conditionally compensate for its absence. We found that L-aspartate semialdehyde sulfurtransferase (L-ASST), catalyzing a sulfurtransferase reaction during homocysteine biosynthesis in methanogens, is potentially involved in genetic compensation of the MMP16 deletion. Even though both L-ASST and MMP16 are members of the COG1900 family, site-directed mutagenesis of conserved cysteine residues implicated in catalysis reveal that the underlying reaction mechanisms may be distinct.

|
Scooped by
?
May 4, 10:54 PM
|
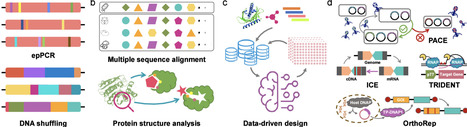
The booming field of synthetic biology and metabolic engineering provides promising approaches for sustainable manufacturing of chemicals from renewable feedstocks with microbial cell factories. Classical metabolic engineering strategies mainly focus on altering gene expression levels and enzyme concentrations to improve the metabolic fluxes of specific pathways. However, the impact and limitations of enzyme properties, which are usually ignored in classical metabolic engineering efforts, can hinder further optimization of microbial cell factories. Protein engineering and directed evolution are powerful tools for modifying proteins to achieve desirable properties, and they have been integrated into metabolic engineering efforts to build highly efficient metabolic pathways and optimal industrial chassis. In this review, we present traditional and data-driven strategies and techniques of directed evolution, including random library design, semirational design, smart library design, and in vivo continuous evolution. We also discuss how these directed evolution strategies have been applied in metabolic engineering toward superphenotypes that cannot be achieved through simple gene overexpression or knockout. Finally, we discuss the challenges of applying protein engineering in metabolic engineering and the prospects for accelerating the directed evolution workflow using the state-of-art technologies.
|
 Your new post is loading...
Your new post is loading...