Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:14 AM
|
A key challenge for antibiotic development against gram-negative bacterial pathogens is their cell biology. These bacteria are enveloped by a second, outer membrane (OM) that has evolved to be an exquisitely selective permeability barrier that can exclude many clinical antibiotics. The OM stands at the frontline in the battle between a pathogen and its host and can come under assault and be damaged. To ensure that OM integrity is continuously maintained, several enzymes act to directly repair and remodel the membrane’s composition and properties. Hererra et al. define a new paradigm for how bacteria can remodel their OM with their discovery of a cell surface–exposed cardiolipin synthase enzyme in Acinetobacter baumannii. This enzyme, ClsO, is remarkable for its unique ability to generate the glycerophospholipid cardiolipin (CL) directly at the bacterial surface. This landmark discovery highlights that our understanding of the ways that diverse bacteria remodel and fortify their OM might only be beginning to scratch the surface.

|
Scooped by
mhryu@live.com
Today, 1:00 AM
|
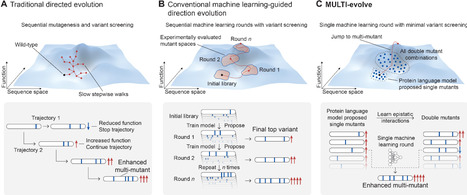
Protein engineering is limited by the inefficient search through a high-dimensional sequence space to find combinations of synergistic mutations. Traditional approaches use stepwise mutation stacking, whereas machine learning methods require extensive datasets or multiple experimental rounds and are bottlenecked by costly, length-limited gene synthesis. We present MULTI-evolve, a rapid evolution framework that systematically engineers multimutants. Our approach combines protein language models or existing functional data with epistatic modelling to predict synergistic combinations. Proposed multimutants are built through MULTI-assembly, a mutagenesis method enabling high-efficiency assembly across multikilobase sequences. Applying MULTI-evolve to three proteins achieved up to 10-fold improvements with a single round of machine learning–guided directed evolution. MULTI-evolve provides a streamlined approach for end-to-end, multimutant engineering for a broad range of protein types and functions.

|
Scooped by
mhryu@live.com
Today, 12:30 AM
|
Coral reefs provide important socioecological services but are vulnerable to climate change, which shifts the balance between the production and erosion of calcium carbonate (CaCO3). In this Review, we summarize understanding of reef accretion, describe the mechanisms of carbonate production and erosion, and consider the effects of future ocean warming and acidification on key reef-building and eroding taxa. The combined stressors of climate change substantially reduce net carbonate production, with a more pronounced effect on calcifying algae than corals. However, declining coral cover driven by marine heatwaves and mass bleaching will probably be the dominant determinant of future reef carbonate budgets, and thus only reefs with thermally adapted populations are predicted to maintain the ability to sustain positive CaCO3 production under climate change, even if calcareous algal cover increases. As carbonate budgets become net negative in the future, the longevity of pre-existing reef frameworks remains unknown and understudied owing to the timescales required to meaningfully assess framework removal rates. Improving estimates of the rates of biologically driven framework loss and chemical dissolution will also be important in better predicting future reef persistence. Key knowledge gaps exist in understanding the effects of deoxygenation on coral reefs, as well as the influence of climate change on understudied sediment-producing taxa such as foraminifera and tropical molluscs. Carbonate production and accretion is negatively affected by ocean warming and acidification, threatening coral reef persistence. This Review synthesizes understanding of environmental impacts on reefs, highlighting the dominant role of heatwaves in reef decline and the importance of thermal adaption for future reef persistence.

|
Scooped by
mhryu@live.com
Today, 12:07 AM
|
Human expansion into deep space necessitates sustainable life support. Current prepackaged food systems face nutritional, logistical, and psychological limits on long missions. Cell agriculture, particularly yeast-based biomanufacturing, emerges as a pivotal solution. Yeast’s genetic flexibility, metabolic resilience, and tolerance to extreme conditions make it an ideal chassis for in-situ food and nutrient production. This review systematically explores yeast’s application in creating closed-loop space food systems, analyzing its progress, challenges, and future potential for enabling sustained extraterrestrial presence.

|
Scooped by
mhryu@live.com
February 22, 12:27 PM
|
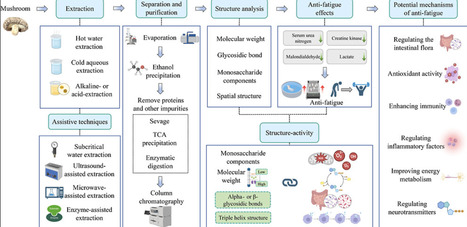
Exercise-induced fatigue is often accompanied by a poor mental state, reduced work efficiency, and decreased athletic performance. Mushroom polysaccharides have been shown in recent years to have a positive effect on alleviating exercise-induced fatigue. This review comprehensively analyzes the extraction methods, structure–activity relationships, and mechanisms of mushroom polysaccharides in alleviating exercise-induced fatigue from 1999 to 2025. Research has revealed that mushroom polysaccharides not only directly alleviate exercise-induced fatigue by regulating gut microbiota but also indirectly mitigate it by influencing oxidative stress, inflammatory factors, and energy metabolism. Moreover, the structure of mushroom polysaccharides is closely related to their biological activity, but there is no systematic consensus on the key structural feature responsible for their antifatigue effects. The purpose of this review is to provide new theoretical evidence for the relationship between the polysaccharide structure and activity and to pave the way for the development of antifatigue functional food ingredients based on mushroom polysaccharides.

|
Scooped by
mhryu@live.com
February 22, 12:21 PM
|
Glycoside hydrolases, “glycosidases”, catalyze carbohydrate catabolism, remodeling, and signaling by accelerating glycosidic-bond cleavage by more than 17 orders of magnitude. Distributed across every kingdom of life and grouped into over 180 sequence-defined families, these enzymes exhibit exceptional diversity in fold, mechanism, and physiological function, and many also catalyze transglycosylation or phosphorolysis. The classical Koshland paradigms─stereochemical inversion, enzymatic nucleophile-assisted retention, and substrate-assisted retention─are analyzed with an emphasis on the conformational itineraries and oxocarbenium ion-like transition states revealed by kinetic isotope effects, linear free-energy relationships, and high-resolution three-dimensional structures. Attention then turns to noncanonical enzymes that employ NAD+-dependent redox hydrolysis or other cleavage mechanisms. Mechanistic insights have inspired the development of engineered glycosidase-derived catalysts for programmed bond construction as well as mechanism-based inhibitors, transition-state analogues, and activity-based probes that are driving advances in chemical biology, biotechnology, and drug discovery.

|
Scooped by
mhryu@live.com
February 22, 12:08 PM
|
E. coli is a well-known host species for studies on recombinant protein expression, typically induced during the exponential phase. Nutrient depletion and the accumulation of toxic substances, which are common during the stationary phase, negatively impact recombinant protein expression, thereby limiting its suitability for efficient protein production. This study aimed to explore a physiological state distinct from conventional strategies for a complementary approach by investigating recombinant protein expression during the stationary phase. An unconventional stationary-like state was engineered using glucose to trigger acetate-mediated growth arrest and reduce tryptone to prevent regrowth. Under these conditions, the absolute and relative expression of signal peptide-fused human interferon gamma (mCsn2-SP-hIFN-γ) increased > 2.4 and 2.5-fold, respectively, compared with that in exponential-phase induction. The present findings suggest that regardless of the unfavorable conditions of the stationary phase for recombinant protein production, glucose utilization may enable the establishment of a stationary-phase model to address these limitations. Overall, these findings underscore the importance of the intracellular metabolic state and expression timing in optimizing recombinant protein production. Although only a single protein was examined in this study, the results highlight the potential of the unconventional stationary phase as an effective new strategy for enhancing recombinant protein expression.

|
Scooped by
mhryu@live.com
February 21, 11:53 PM
|
Bacterial conjugation is a major route of horizontal gene transfer and a key driver of the dissemination of antibiotic resistance, exacerbating the global health crisis. Central to this process is TraI, a bifunctional relaxase-helicase encoded by the iconic F-plasmid family. TraI initiates single-stranded DNA (ssDNA) transfer by covalently attaching to plasmid DNA through its trans-esterase (TE) activity and subsequently unwinds the plasmid via its helicase activity. Although genetic and biochemical studies have postulated that two TraI molecules are required for efficient conjugative transfer, how these molecules coordinate their activities in space and time - and how strand nicking is coupled to helicase loading - has remained unknown. Here, we capture the elusive, functionally asymmetric TraI homodimer and report its cryo-EM structure, revealing that the interaction between the two TraI molecules is mediated predominantly by ssDNA. We further show that TE-driven duplex melting at the origin of transfer (oriT) by one TraI molecule enables helicase loading by a second TraI molecule on the transfer strand of the unnicked plasmid DNA. Although the TE domain is intrinsically competent for strand nicking, its activity is inhibited by the host factor IHF, and this inhibition is relieved upon helicase loading. Together, these findings define a regulatory loop that coordinates helicase loading with strand nicking, providing mechanistic insight into the earliest stages of conjugative DNA processing.

|
Scooped by
mhryu@live.com
February 21, 2:05 PM
|
Periodontitis is a chronic inflammatory disease driven by subgingival dysbiosis. Despite progress in characterizing microbial diversity, functional heterogeneity at the single-cell level remains poorly understood, mainly due to challenges from low microbial biomass and host contamination. Here, we present a single-cell RNA sequencing framework for profiling subgingival bacteria, enabling high-resolution analysis of these communities in health and periodontitis. Using 16 subgingival samples, we generated an atlas spanning 133,458 cells across 285 species, including 57 core active species grouped into eight functional clusters. Health-associated subpopulations specializing in adhesion and polysaccharide degradation decline in periodontitis, coinciding with hypoxia and elevated amino acid availability. The keystone pathogens T. denticola and P. gingivalis display species-specific tendencies in amino acid metabolism-related transcriptional profiles, while P. intermedia harbors a proteolytic subpopulation enriched in periodontitis. These insights deepen our understanding of periodontitis pathogenesis and inform precision diagnostics and therapeutic strategies.

|
Scooped by
mhryu@live.com
February 21, 1:11 PM
|
Advancements in synthetic genetic circuits have enabled programmable and condition-dependent control of microbial cell growth. CRISPR-Cas9-based kill switches, genetic systems that program cells to lose viability in response to specific conditions, have recently been demonstrated for bacterial cell factories but not yet in yeast. Here, we present a foundational demonstration for a CRISPR-based kill switch in Saccharomyces cerevisiae, CRISPR KiSS. The CRISPR KiSS employs inducible CRISPR targeting essential genes to elicit growth inhibition. The activation of the KiSS system is achieved through conditional expression of a guide RNA (gRNA) upon anhydrotetracycline (ATc) induction, thereby activating CRISPR-mediated gene disruption. We demonstrate that targeting the essential genes (ERG13, PGA3, TPI1 or CDC19) leads to severe growth inhibition upon ATc induction. Still, the current set up does not allow complete killing of the cells due to system inactivation, e.g. escape from CRISPR based cutting. We studied reasons for system inactivation and substantially improved the system by simultaneous expression of two different gRNAs. Sequencing escape mutants revealed mutations in both the gRNA sequences and target genes as potential sources of system inactivation. This work highlights the potential of harnessing a CRISPR-based kill switch in S. cerevisiae. Cells expressing the system were able to escape growth inhibition through mutations and further optimization of the KiSS system is still needed for it to be used in various cell factory applications.

|
Scooped by
mhryu@live.com
February 21, 1:05 PM
|
Phages are a valuable resource for the genetic engineering of Streptomyces antibiotic-producing bacteria. Indeed, a few integrative vectors based on phage integrase are available to insert transgenes at specific genomic loci. Chromosome conformation captures previously demonstrated that the Streptomyces linear chromosome is organized in two spatial compartments: the central compartment encompassing the most conserved and highly expressed genes in exponential phase, and the terminal compartments enriched in poorly conserved sequences including specialized metabolite biosynthetic gene clusters. This study introduces a new integrative tool based on a recently described phage, Samy, which specifically targets the terminal compartment of its native host chromosome. Samy is related to PhiC31 phage and, like the latter, encodes a serine integrase. Whereas PhiC31 targets a site generally located near the origin of replication, the Samy integration site is one of the farthest known attB sites from it. We demonstrated that the Samy integrase efficiently mediates the specific integration of a non-replicating plasmid in six Streptomyces strains from distinct clades. Bioinformatic analyses revealed that the Samy-attB site is rather conserved and located in the terminal compartment of most Streptomyces chromosomes. Finally, heterologous expression of the albonoursin biosynthetic gene cluster from the Samy-, PhiC31-, and R4-attB sites yields quantitatively equivalent levels of production, though qualitative differences were observed. Altogether, these results demonstrate that the att-int Samy system expands Streptomyces genetic engineering tools by enabling targeted integration in the terminal chromosomal compartment.

|
Scooped by
mhryu@live.com
February 21, 12:56 PM
|
While extracellular vesicles (EVs) are established mediators of intra-species signaling, their contribution to cross-kingdom communication remains incompletely understood. Here, we investigate the EV-mediated interactions between human colon epithelial cells and both Gram-positive and Gram-negative gut bacteria. We show that bacterial EVs (BEVs) derived from Lacticaseibacillus casei, Enterococcus faecalis, and Proteus mirabilis induce distinct transcriptomic changes in Caco-2 cells depending on the bacterial species, with up to ~6,000 differentially expressed genes, including CCL20, CXCL8, or CXCL10. Transfection of BEV-derived RNA independently induces a subset of similar effects, indicating that the EV-mediated communication is partially driven by the RNA cargo. Conversely, we demonstrate that bacteria interact with Caco-2-derived EVs and miR-192-5p, which is highly abundant (~36.4-fold higher) in EVs isolated from conditioned medium compared with EVs from unconditioned medium, with modest effects on bacterial growth. Furthermore, we show that lipid-based packaging of miR-192-5p modulates its association with the bacteria. Our findings support a conceptual model in which EVs and their RNA cargo contribute to species-dependent host-microbe interactions. This study introduces a framework for understanding EVs as cross-kingdom regulators and underscores the importance of tailored, context-specific analyses for understanding the scope of EV-mediated interactions in microbiome-host homeostasis and disease.

|
Scooped by
mhryu@live.com
February 21, 12:30 PM
|
Cyanobacteria are known for their rich secondary metabolome with a long history of research directed toward the industrial and pharmaceutical applications of their natural products. Cyanobacterial metallophores (metal-chelating molecules), however, are understudied relative to metallophores from other phyla despite evidence suggesting that genes for metallophore biosynthesis are well-represented in cyanobacterial genomes. Many of the characterized cyanobacterial metallophores are formed from hybrid biosynthetic pathways and feature mixed coordinating functional groups, leading to enhanced structural and functional diversity. The few characterized metallophore families have intriguing properties including promiscuity of metal binding, photoreactivity, and amphiphilicity that are yet to be fully explored. Research suggests that these compounds are ecologically relevant and could guide community dynamics by controlling the availability of iron, detoxifying copper, and allelopathically inhibiting certain organisms. Cyanobacterial metallophores also have potential in the fields of therapeutic design, bioremediation, technology, and agriculture. In the past five years, the number of characterized metallophores from cyanobacteria has doubled, pointing to the great promise for future discoveries.
|

|
Scooped by
mhryu@live.com
Today, 1:08 AM
|
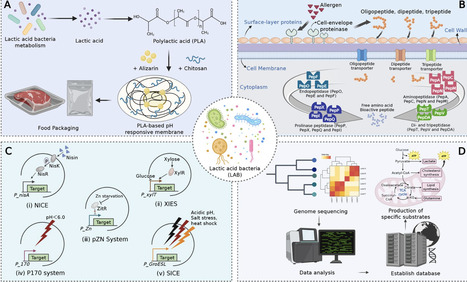
Lactic acid bacteria (LAB) have transitioned from their traditional roles in food preservation and flavor enhancement to more advanced applications supported by synthetic biology, genome editing, and artificial intelligence (AI). This review provides a comprehensive overview of how LAB contribute to improvements in the quality of fermented foods, including their texture, flavor, and nutritional benefits. It evaluates innovative strategies such as CRISPR-based strain improvements, microfluidic high-throughput screening, AI-enhanced precision design, and personalized nutrition frameworks that connect microbiome profiles to specific LAB functionalities. Additionally, it discusses significant challenges, including global regulatory differences, ethical concerns regarding genetically engineered LAB, technical issues related to scaling advanced fermentation technologies, and consumer acceptance. Overcoming these barriers requires a coordinated, interdisciplinary approach. Therefore, future research should focus on integrating multi-omics platforms, establishing standardized LAB functional databases, and developing AI-driven precision nutrition tools to promote sustainable and consumer-friendly innovation in the fermented food industry.

|
Scooped by
mhryu@live.com
Today, 12:50 AM
|
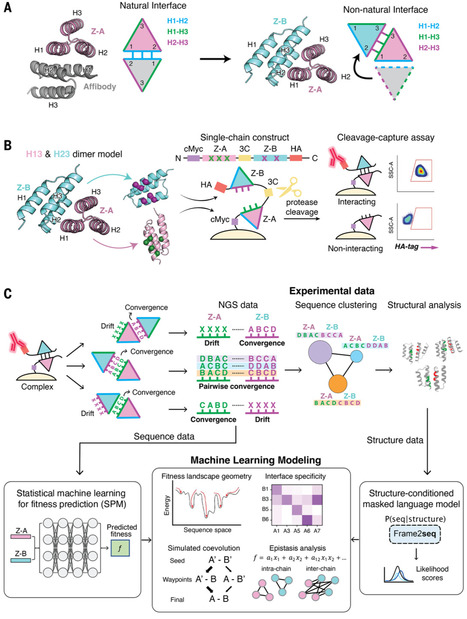
Understanding how protein binding sites evolve interactions with other proteins could hold clues to targeting “undruggable” surfaces. We used synthetic coevolution to engineer new interactions between naïve surfaces, simulating the de novo formation of protein complexes. We isolated seven distinct structural families of protein Z-domain complexes and found that synthetic complexes explore multiple shallow energy wells through ratchet-like docking modes, whereas complexes formed by natural binding sites converged in a deep energy well with a relatively fixed geometry. Epistasis analysis of a machine learning–estimated fitness landscape revealed “seed” contacts between binding partners that anchored the earliest stages of encounter complex formation. Our results suggest that “silent” surfaces have a shallower energy landscape than natural binding sites, disfavoring tight binding, likely owing to evolutionary counterselection.

|
Scooped by
mhryu@live.com
Today, 12:11 AM
|
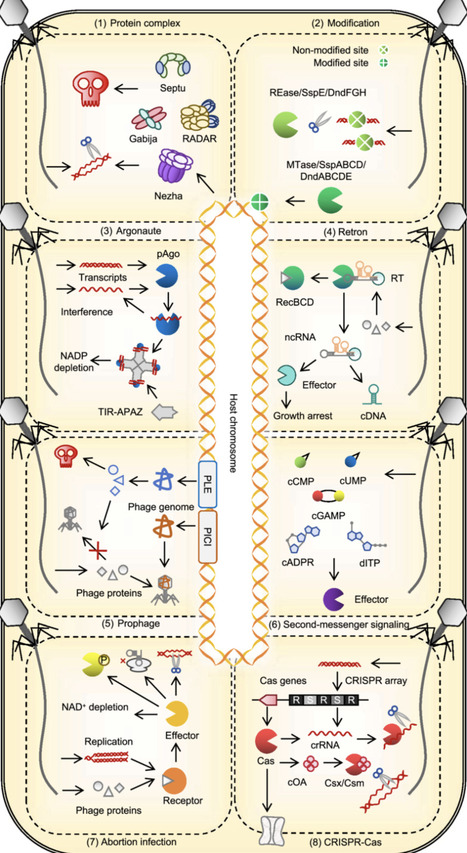
Bacteriophages and archaeal viruses are the most abundant biological entities on Earth. Through a long-standing co-evolutionary arms race, they have driven the emergence of a diverse repertoire of prokaryotic defense systems. This review summarizes these systems, highlighting their diverse antiviral mechanisms across distinct stages of viral infection, from surface barriers and inducible innate responses to specific adaptive defenses, and the intricate interplay between these defense strategies. By examining host–virus counter defense dynamics, the trade-off between survival benefit and adaptive cost, the co-evolution of RNA and protein components, and the comparison with eukaryotic immune systems, we underscore the intrinsic complexity and evolutionary plasticity of prokaryotic antiviral immunity. A deeper understanding of these processes and mechanisms will not only shed light on the origins and evolution of the immune system but also provide valuable opportunities for the development of biotechnological tools.

|
Scooped by
mhryu@live.com
Today, 12:02 AM
|
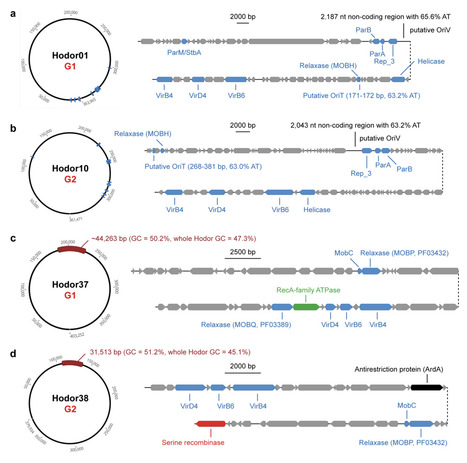
Closely related prophages are frequently found in phylogenetically distant bacteria in the human gut, despite limited evidence of productive phage infections across broad host ranges. Thus, it remains unclear how the wide distribution of prophages could emerge. Here, we identify a potential mechanism of prophage dissemination. We describe two deeply diverged groups of conjugative megaplasmids (>300 kilobases) in the human gut microbiome, which we term Hodors. Hodors encode conserved replication, partitioning, and type IV secretion systems, together with a complex surface-associated gene module. A subset of Hodors harbor complete, intact prophage genomes, and closely related prophages are detected across phylogenetically distant Bacillota lineages, including both Bacilli and Clostridia. Further analysis indicates that Hodor-associated prophages can exist as extracellular particles and demonstrate their transcriptional activity. Our findings support a model in which conjugative megaplasmids act as composite mobile platforms that disseminate prophage genomes across bacterial lineages, providing a mechanistic explanation for the widespread occurrence of closely related prophages in phylogenetically distant gut bacteria and effectively decoupling lysogenic host range from infective host range.

|
Scooped by
mhryu@live.com
February 22, 12:24 PM
|
Using cellulase to degrade cellulose for cellobiose production is an economically competitive pathway for the high-value utilization of lignocellulose. Cellobiohydrolases hydrolyze cellulose into cellobiose but suffer severe product inhibition, whereas endoglucanases (EGs) randomly cleave cellulose chains into oligosaccharides. Three heterologously expressed EGs from different sources (PcCel45A, BsCel5, and GtCel5A) were screened for their ability to degrade cellulose into cellobiose, and their hydrolysis processes were characterized. Using PcCel45A and BsCel5 to hydrolyze Avicel step-by-step at a protein ratio of 1:1 produced cellobiose with a purity of 90.99% and a yield of 1.2 g per 100 g of Avicel. Further studies revealed that PcCel45A and BsCel5 cannot degrade cellobiose, and their activities are not inhibited by the product, showing their advantage in cellobiose production. Additionally, adsorption kinetics and thermodynamic analyses indicated that PcCel45A and BsCel5 exhibited distinct adsorption mechanisms. This study provides a promising new strategy for the high-value utilization of lignocellulose.

|
Scooped by
mhryu@live.com
February 22, 12:15 PM
|
High-yielding recombinant protein expression systems often face challenges due to the metabolic burden caused by the competition for cellular resources, resulting in reduced growth and, hence, limiting their industrial applicability. Furthermore, industrial recombinant protein production is also affected by the occurrence of oxygen gradients, which is a prevalent issue in large-scale bioreactors. These gradients create a heterogeneous environment in the bioreactor, which affects cell growth and metabolism, having severe consequences on the process performance. Both these factors alter cellular physiology and metabolism, thereby affecting recombinant protein yields. Understanding metabolic adaptations to these stress conditions is crucial for uncovering the underlying cellular mechanisms, which can direct further optimization of the recombinant strains. In this study, we aimed to explore the combined response of the central metabolism of E. coli to metabolic burden and microaerobic conditions. Two recombinant protein-producing E. coli BL21 strains carrying XylS/Pm vectors with low (A2-mCh) and medium plasmid copy numbers (A3-mCh), and producing mCherry protein, were studied by introducing oxygen limitation. Central metabolite pools were analyzed by three targeted LC–MS/MS methods, using the isotope dilution strategy for absolute quantification. Both recombinant strains exhibited different levels of metabolic burden, with the strain possessing a higher plasmid copy number showing more pronounced growth retardation and a stronger impact on metabolite pools. Both strains, however, showed a similar response to oxygen limitation, with significant adaptations in the central metabolite pools. The low plasmid copy number strain showed an increase in the concentration of lower glycolytic and tricarboxylic acid cycle metabolites, while the pools of upper glycolytic and pentose phosphate pathways and nucleoside phosphates were mostly unaffected. However, a more extreme response was seen in A3-mCh, where the majority of the metabolite pools were increased. Oxygen limitation caused lower metabolic activity, but the energy charge and redox balance were maintained, and no negative effect was observed on mCherry production rates. This study provides insights into metabolic adaptations in E. coli BL21 recombinant strains, having quite robust mechanisms to maintain intracellular metabolic homeostasis during both internal and external perturbations.

|
Scooped by
mhryu@live.com
February 22, 11:48 AM
|
Anthropogenic nitrogen enrichment is widely expected to suppress symbiotic nitrogen fixation in terrestrial ecosystems. Nevertheless, observed symbiotic nitrogen fixation responses remain incompletely explained by exogenous nitrogen inputs, climate, and edaphic factors. In this meta-analysis, we integrate 908 globally distributed field measurements to identify the key predictors that improve simulation of symbiotic nitrogen fixation responses to nitrogen enrichment. On average, symbiotic nitrogen fixation declines by 33.0% upon nitrogen enrichment, with the reduction being more pronounced in non-croplands than croplands. Models considering only environmental factors overestimate symbiotic nitrogen fixation decline relative to observations. The better performance of plant traits like plant growth and biomass allocation (shoot:root ratio) partially buffer symbiotic nitrogen fixation suppression under nitrogen enrichment. Integrating both environmental factors and plant performance traits improves predictive accuracy of symbiotic nitrogen fixation responses by 42.7% and brings the simulated symbiotic nitrogen fixation reductions into close agreement with observations. The alterations in plant performance traits are thus critical for explaining variability in terrestrial symbiotic nitrogen fixation responses, and incorporating plant trait dynamics in Earth System Models can quantitatively partition the compensatory symbiotic nitrogen fixation supported by nitrogen-fixing plant growth from the direct negative impact of nitrogen inputs. Anthropogenic nitrogen enrichment reduces symbiotic nitrogen fixation, but environmental factors alone cannot explain this response. This meta-analysis shows that integrating both environmental factors and plant performance traits improves the predictive accuracy of symbiotic nitrogen fixation responses by 42.7%.

|
Scooped by
mhryu@live.com
February 21, 2:11 PM
|
Natural products are vital sources of medicines, and their analogues hold great promise as novel therapeutics to combat disease more effectively, including circumventing antibiotic resistance. Engineering biosynthetic pathways in microbial hosts enables efficient, cost-effective production of non-natural analogues — compounds structurally related to natural products but not necessarily derived through direct chemical modification. In this review, we highlight recent work on the in vivo production of novel natural product analogues, with an emphasis on combinatorial and precursor-directed biosynthesis, enzyme engineering, and retrobiosynthesis. We anticipate that further developments in artificial intelligence, particularly the use of machine learning models to understand enzymatic transformations and predict novel reactions, will significantly accelerate this field and drive forward its importance in drug discovery and related research.

|
Scooped by
mhryu@live.com
February 21, 1:33 PM
|
Antibiotic resistance is a growing global health threat. Although antibiotic activity is well studied in homogeneous liquid cultures, many infections are caused by spatially structured multicellular populations where consumption of scarce nutrients establishes strong spatial variations in their abundance. These nutrient variations have long been hypothesized to help bacterial populations tolerate antibiotics, since liquid culture studies link antibiotic tolerance to metabolic activity, and thus, local nutrient availability. Here, we test this hypothesis by visualizing cell death in structured E. coli populations exposed to select nutrients and antibiotics. We find that nutrient availability acts as a bottleneck to antibiotic killing, causing death to propagate through the population as a traveling front. By integrating our measurements with biophysical theory and simulations, we establish quantitative principles that explain how collective nutrient consumption can limit the progression of this “death front,” protecting a population from a nominally deadly antibiotic dose. While increasing nutrient supply can overcome this bottleneck, in some cases, excess nutrient unexpectedly promotes the regrowth of resistant cells. Altogether, this work provides a key step toward predicting and controlling antibiotic treatment of spatially structured bacterial populations, yielding biophysical insights into collective behavior and guiding strategies for effective antibiotic stewardship. In this work, authors provide evidence that bacteria in spatially structured populations protect each other from antibiotics through collective nutrient consumption, creating ‘death fronts’ that sweep through the colony– explaining why infections often survive treatments that work in lab tests.

|
Scooped by
mhryu@live.com
February 21, 1:08 PM
|
With antimicrobial resistance emerging as a top global health concern, new approaches to prevent and treat infections are urgently needed. Antimicrobial peptides are recognized as a promising alternative to conventional antibiotics. Considering the costly, labor-intensive, and time-consuming nature of wet lab screening, in recent years, proteomics research has increasingly relied on in silico methods to predict physicochemical, structural, and biological properties of candidate peptides. Here, we present the AMPSeek protocol to enable scientists to screen peptides and predict their potential antimicrobial activity, toxicity, and associated three-dimensional structure with a single command. The AMPSeek framework integrates AMPlify, tAMPer, and LocalColabFold, scales efficiently with increasing sample size, and produces an interactive HyperText Markup Language report to facilitate result interpretation. We provide step-by-step instructions for installing and running AMPSeek on a ten-peptide test set, along with two additional datasets (n = 20 and n = 100; mean peptide length ≈121.3) to demonstrate AMPSeek's near-linear scalability.

|
Scooped by
mhryu@live.com
February 21, 1:01 PM
|
The transition to low-carbon energy systems requires scalable and energy-efficient routes for producing liquid biofuels that are compatible with existing fuel infrastructures. This review focuses on bio-oil production from phototrophic microorganisms, highlighting their high biomass productivity, rapid growth, and inherent capacity for carbon dioxide fixation as key advantages over conventional biofuel feedstocks. Recent progress in thermochemical conversion technologies, particularly hydrothermal liquefaction (HTL) and fast pyrolysis, is critically assessed with respect to their suitability for wet and dry algal biomass, respectively. HTL enables direct processing of high-moisture biomass while avoiding energy-intensive drying, whereas fast pyrolysis offers high bio-oil yields from lipid-rich feedstocks. In parallel, catalytic upgrading strategies, including hydrodeoxygenation and related hydroprocessing routes, are discussed as essential steps for improving bio-oil stability, heating value, and fuel compatibility. Beyond conversion technologies, innovative biological and biotechnological strategies, such as strain optimization, stress induction, co-cultivation, and synthetic biology approaches, are examined for their role in tailoring biomass composition and enhancing bio-oil precursors. The integration of microalgal cultivation with wastewater utilization is briefly considered as a supporting strategy to reduce production costs and improve overall sustainability. Overall, this review emphasizes that the effective coupling of advanced thermochemical conversion with targeted biological optimization represents the most promising pathway for scalable bio-oil production from phototrophic microorganisms, positioning algal bio-oil as a viable contributor to future low-carbon energy systems.

|
Scooped by
mhryu@live.com
February 21, 12:50 PM
|
In Azotobacter vinelandii, the sigma factor RpoS is maintained at low levels in exponentially growing cells due to degradation mediated by the chaperone–protease complex ClpXP, while high levels are observed in the stationary phase. This study showed that degradation of RpoS by ClpXP is under the control of the Gac-Rsm signal transduction system, in which GacA, the transcriptional activator of the two-component system GacS/GacA, activates transcription of the small RNAs RsmZ1 and RsmZ2. These RNAs bind to the translational repressor protein RsmA to counteract its repressor activity on its target mRNAs. We found that in stationary-phase cells, compared with the WT, a gacA mutant exhibited lower RpoS levels due to reduced stability, while levels of the clpP and clpX mRNAs were higher. Furthermore, inactivation of the clpP or clpX genes in the gacA mutant restored the stability of RpoS, suggesting that the observed RpoS instability is due to degradation by ClpXP. We also showed that inactivation of rsmA in either the WT or the gacA mutant resulted in RpoS levels higher than in the WT in both stationary and exponential phases, while clpP and clpX transcript levels were significantly reduced. Taken together, these data reveal that in A. vinelandii, the GacA-Rsm system controls RpoS stability through RsmA, which acts as a positive regulator of ClpXP expression.
|
 Your new post is loading...
Your new post is loading...