 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 5:09 PM
|
Horizontal gene transfer (HGT) is a promising avenue for microbiome engineering that enables DNA delivery to microbes in their native environment. Integrative and conjugative elements (ICEs), a class of genomically-integrated mobile elements, are ideal vectors for this purpose. Developing targeted ICE transfer for controlled microbiome engineering requires a better understanding of ICE mobility in complex communities. However, current methods for tracking horizontal gene transfer are not high throughput. To improve upon current methods, we developed a sequencing-based strategy to track and quantify ICE transfer in complex mixtures that we refer to as ‘ICE-seq.’ This method was able to assess ICE transfer in a synthetic community of 150 recipients simultaneously and in combination with phenotypic assays demonstrated that restriction-modification (RM) systems in the donor and recipient alter ICE transfer rate with subspecies resolution by up to 10,000-fold. We propose that manipulating RM systems can be a strategy to engineer targeted ICE transfer for directed microbiome engineering.

|
Scooped by
mhryu@live.com
Today, 5:01 PM
|
Isoquercetin is a bioactive flavonoid whose biosynthesis from quercetin depends on glycosyltransferases (GTs); however, wild-type GTs exhibit a limited catalytic efficiency. Here, we combined virtual screening, molecular docking, and experimental validation to identify and engineer the GTs. From about 1000 homologous sequences screened via protein BLAST and deep learning-based kcat prediction, the glycosyltransferase MiCGT from Mangifera indica was selected for rational design. After molecular docking and phylogenetic analysis, the engineered variant PCAA (MiCGTS121P/M148C/K253A/S281A) exhibited a 103-fold higher activity than wild-type MiCGT. To enable cost-effective production, sucrose synthase GmSUS was coupled with PCAA to regenerate UDP-glucose during quercetin glycosylation, yielding 3.91 mM isoquercetin with a 78.1% conversion rate. This work overcomes a major bottleneck in isoquercetin biosynthesis and offers a practical strategy for its application in the food industry.

|
Scooped by
mhryu@live.com
Today, 4:47 PM
|
Gut microbe cultivation is essential for studying host-microbiota interactions. Traditional cultivation methods often fail to recover microbial species at low abundance (< 0.1%). To overcome this limitation, we employed the bent-capillary-centrifugal-driven (BCCD) method to encapsulate and cultivate fecal microbes in microdroplets. Fecal bacterial cells were distributed into ~50 nL microdroplets via the BCCD generator, and the microdroplets were dispersed in the oil phase and further incubated under controlled conditions. The BCCD method significantly increased the frequency of microbes at low abundance. Compared to the plate-based method, BCCD-based cultivation produced distinct microbial community structures and exhibited significantly lower temporal variation during cultivation (p < 0.05). Lineage-specific effect size (LEfSe) analysis revealed that BCCD-based cultivation enriched 29 low-abundant bacterial genera, whereas the plate-based method enriched 26. Using this method, we isolated 1,049 bacterial strains representing 123 species and 58 genera, including 8 novel species. Among the isolated and cultivated genera, 62.1% (36/58) were microbes of low abundance in the original fecal sample, and 41.4% (12/29) of the BCCD-specific enriched genera were successfully obtained. Notably, comparison with four major gut microbial culture studies revealed 45 species were exclusively recovered in this work. Taken together, the results demonstrated that our BCCD-based cultivation method effectively enriched and facilitated the isolation and cultivation of microbes at low abundance and novel gut bacterial species.

|
Scooped by
mhryu@live.com
Today, 4:35 PM
|
The microbiota–gut–brain axis, defined as the bidirectional communication linking the gut microbiota with the brain, operates through neural, metabolic, immune, and endocrine signals. It plays an important role in mental health, regulating stress response, mood, and cognitive function, and is altered in psychiatric conditions. Although the gut microbiota remains stable during healthy adulthood, it is modifiable by lifestyle, medication, and diet, making it a tractable target for mental health interventions. Diet is emerging as a viable option to improve mental health through gut microbiota modulation. Energy-dense, high-fat, and high-sugar diets have been linked to poor mental health, whereas Mediterranean, fiber-rich, and fermented-food diets show benefits, possibly through provision of specific nutrients and beneficial microbial metabolites such as short-chain fatty acids. However, more human research addressing variability and confounding is needed to unlock these mechanisms and support personalized/precision nutrition. This review discusses current evidence and proposes multidisciplinary, rigorous diet–microbiota–mental health research.

|
Scooped by
mhryu@live.com
Today, 4:07 PM
|
Protein C-termini can vary due to errors or programmed regulation, contributing to proteome diversity, yet their impact on the proteome remains poorly understood. Although aberrant C-termini are often linked to protein degradation, it is unclear if this holds true universally. In this study, we examine how C-terminal variations—arising from disease-associated nonstop mutations, alternative splicing, and translational readthrough—affect protein half-lives. Our findings indicate that, contrary to previous studies, erroneous C-termini can either stabilize or destabilize proteins. We have identified multiple oncoproteins and tumor suppressors whose protein stability is altered by disease-relevant nonstop mutations. Notably, we have found that C-terminal variations commonly influence the stability of canonical proteins, extending beyond their role in protein quality control. Furthermore, we have uncovered C-terminal features that distinguish erroneous from wild-type proteins and reveal that hydrophobic C-termini are targeted by a complex ubiquitin ligase network. Overall, our work broadens the understanding of C-terminal-dependent protein degradation and supports that C-terminal variation is a widespread strategy for generating protein forms with distinct half-lives to exert diverse biological functions. Protein C-terminal diversity is widespread, yet its proteome-wide impact remains unclear. Here, the authors show that C-terminal variations influence the stability of canonical and disease associated mutant proteins and define the sequence features and degradative mechanisms underlying this regulation.

|
Scooped by
mhryu@live.com
Today, 3:47 PM
|
The regulation of redox balance and energy conservation is fundamental to life and relies on a large evolutionary network of oxidoreductases forming homologous protein complexes, collectively termed HORBEC (homologous oxidoreductase complexes involved in redox balance and energy conservation). These include hydrogenases, respiratory complex I and electron-bifurcating complexes, central to respiration, fermentation and methanogenesis. Despite their crucial role, a comprehensive investigation of the diversity and evolutionary history of HORBEC has been lacking. Here we exhaustively identified and analysed over 50 protein families representing all HORBEC components across thousands of bacterial and archaeal genomes. We propose a unified nomenclature and classification encompassing 31 complexes and provide an annotation tool. We highlight the extensive diversity of HORBEC, especially in Archaea. We provide information on overlooked systems and identify a new one probably acting as a cation transport platform. We show that HORBEC originated via extensive tinkering of ancestral modules, driven by strong evolutionary constraints. Finally, we infer the presence of respiratory complex I in the last universal common ancestor, opening questions on its potential role in early energy metabolisms. This work provides an evolutionary framework for HORBEC, representing a fundamental resource to predict and study redox metabolisms of ecological and biotechnological significance. HORBEC are protein complexes involved in the regulation of redox balance and energy conservation. The authors develop a bioinformatic tool for HORBEC annotation in bacterial and archaeal genomes and reconstruct the evolutionary history of these fundamental enzymes.

|
Scooped by
mhryu@live.com
Today, 3:30 PM
|
Although endophytic microorganisms play a critical role in plant growth and stress resilience, the genetic basis underlying host selection of beneficial microbiota—particularly within the xylem—remains poorly understood. Cucumber (Cucumis sativus), as a crop model with a well-developed system for studying vascular biology, offers a valuable system to investigate the host genetic determinants of xylem microbiome assembly. By conducting population-level microbiome profiling across 109 cucumber accessions, we identified a conserved xylem microbiota dominated by Proteobacteria. Within this community, 20 core amplicon sequence variants (ASVs) were consistently present in xylem sap. Genome-wide association mapping identified a host genetic locus, CsXPR1, which encodes a tetratricopeptide repeat protein that regulates the abundance of the dominant xylem-colonized Pseudomonas ASV_4. Colonization patterns of ASV_4 varied across host genotypes and were correlated with CsXPR1 expression levels, suggesting a precision genetic regulation of bacterial entry into vascular tissues. Pseudomonas fulva strain 220, with 97% 16S rRNA gene identity with ASV_4, could colonize in cucumber xylem by inoculation of either roots or leaves. Genome analysis and plate assays revealed the biosynthesis of indole-3-acetic acid (IAA), solubilization of phosphate, and a range of plant beneficial traits in strain 220. Inoculation with strain 220 significantly enhanced growth in cucumber, but only in CsXPR1 haplotype that exhibited high gene expression and higher recruitment capacity of the strain. These benefits included notable increases in plant height (38%), stem diameter (36%), leaf area (61%), fresh and dry weight (51% and 85%, respectively), and a 4.57-fold increase in 4-methyleneglutamine content within the xylem sap. Our findings reveal a complete “gene-to-function” pathway where the host gene CsXPR1 mediates a genotype-dependent growth promotion. It achieves this by regulating the xylem colonization of a beneficial bacterium, Pseudomonas fulva, which in turn enhances plant growth by enriching the xylem sap with the key metabolite 4-methyleneglutamine.

|
Scooped by
mhryu@live.com
Today, 3:07 PM
|
RNA polymerase (RNAP) must unwind duplex DNA prior to transcription initiation. In bacteria, unwinding starts at a DNA motif called the promoter −10 element. Specifically, non-template strand bases interact with RNAP to trigger the process. A protein called CarD can support −10 element opening in many microbes. Whilst this most often activates transcription, CarD may repress if the DNA open complex is too stable. For Rhodobacter sphaeroides, a purple photoheterotrophic alpha-proteobacterium, CarD is particularly important as many −10 elements have a key DNA sequence defect. Here, we use genomic tools to map transcription initiation, binding of RNAP, CarD interactions, and DNA topology, globally in R. sphaeroides. We show an association between CarD-controlled transcription, and negative DNA supercoiling, which alters global gene expression if perturbed. Using biochemical tools, we show that promoter co-regulation by CarD and supercoiling results from the −10 element defect inherent to R. sphaeroides promoters. If this flaw is corrected, regulation by CarD and supercoiling is disrupted. As supercoiling dissipates during stress, we suggest CarD couples housekeeping transcription to the environment via DNA topology. The transcription factor CarD facilitates the activation of transcription in many bacteria and in Rhodobacter sphaeroides, CarD compensates for suboptimal promoter DNA sequences. Here, the authors show CarD function is tied to promoter DNA topology.

|
Scooped by
mhryu@live.com
Today, 11:30 AM
|
Argonautes are an evolutionarily conserved family of proteins that use guide oligonucleotides for specific recognition of nucleic acid targets. While eukaryotic Argonautes share a conserved structure and act in RNA interference, prokaryotic Argonautes (pAgos) display remarkable structural and functional variations, including various guide and target specificities. Most studied catalytically active pAgos use 5′-phosphorylated DNA guides to recognize and cleave DNA targets. Here, we describe a new group of pAgos from mesophilic bacteria that use RNA guides to cleave DNA targets and are active at physiological temperatures. In contrast to most characterized pAgo nucleases, these proteins can utilize guides of varying lengths containing either a 5′-phosphate or a 5′-hydroxyl group with similar efficiencies. Measurements of the kinetics of target DNA binding show that the annealing of the guide–target duplex in pAgo occurs in an ordered way and that pAgo increases the rate of guide–target interaction in the seed region. We show that the analyzed pAgos can produce alternative DNA products depending on the structure of guide RNA and the cleavage conditions. The results demonstrate that RNA-guided pAgo nucleases are more widespread than previously anticipated and that these pAgos have a relaxed specificity for target DNA cleavage.

|
Scooped by
mhryu@live.com
Today, 11:14 AM
|
Structural-Maintenance-of-Chromosome (SMC) complexes, such as condensins, organize the folding of chromosomes. However, their role in modulating the entanglement of DNA and chromatin is not fully understood. To address this question, we perform single-molecule and bulk characterization of yeast condensin in entangled DNA. First, we discover that yeast condensin can proficiently bind double-stranded DNA through its hinge domain, in addition to its heads. Through bulk microrheology assays, we then discover that physiological concentrations of yeast condensin increase both the viscosity and elasticity of dense solutions of -DNA, suggesting that condensin acts as a crosslinker in entangled DNA, stabilising entanglements rather than resolving them and contrasting the popular theoretical picture where SMCs purely drive the formation of segregated, bottle-brush-like chromosome structures. We further discover that the presence of ATP fluidifies the solution–likely by activating loop extrusion–but does not recover the viscosity measured in the absence of protein. Finally, we show that the observed rheology can be understood by modelling SMCs as transient crosslinkers in bottle-brush-like entangled polymers. Our findings help us to understand how SMCs affect the dynamics and entanglement of genomes.

|
Scooped by
mhryu@live.com
Today, 1:58 AM
|
There is still a need for a better understanding of how abiotic/biotic factors affect the functional structure and composition of biological assemblages, given that living organisms are in constant interaction with their environment and with each other. Here, we present a comprehensive dataset of 31 functional traits of bacteria using information from BacDive, a bacterial diversity meta-database, as well as from the rrnDB and genomesizeR datasets. This updated version of the BactoTraits dataset, in addition to now offering more traits for more strains (97,721 strains with at least one trait described), makes R scripts available to the scientific community. These traits include physiological characteristics, metabolic processes, genome properties and biotope preferences. They could be inferred to the whole bacterial community thanks to taxonomic affiliation obtained from traditional high throughput 16S rRNA gene amplicon sequencing methods. This taxonomic affiliation is based on the regularly updated SILVA database and thus allows to study combinations of weighted mean trait profiles of bacterial communities at different taxonomic levels. BactoTraits can be used, for example, to improve predictions of ecological responses to natural/anthropogenic pressures and to support biomonitoring, management and conservation strategies. The R scripts, as well as the dataset encoded in BactoTraits, are available at: https://doi.org/10.24396/ORDAR-182.

|
Scooped by
mhryu@live.com
Today, 1:45 AM
|
Plants and microbes exchange macromolecules such as RNA and proteins. How this exchange is accomplished is poorly understood, but extracellular vesicles (EVs) have been proposed as likely vehicles. Here, we review recent work on the biogenesis and functions of plant EVs and the current evidence in support of and against their role in cross-kingdom RNA interference. Plant EVs, like EVs from other kingdoms of life, are released in part by the fusion of multivesicular bodies with the plasma membrane, a complex and conserved mechanism involving lipid-modifying proteins, the exocyst complex, and Rab GTPases. Though some plant EV subpopulations are involved in immunity, it appears unlikely that plant EVs contribute to cross-kingdom RNA interference. Recent work has shown that plants secrete extravesicular RNA, including small RNAs and long noncoding RNAs, into the leaf apoplast and onto leaf surfaces, while very little RNA is found inside of EVs. We propose that these free extracellular RNAs play a central role in maintaining a healthy leaf microbiome.

|
Scooped by
mhryu@live.com
Today, 1:29 AM
|
Homomeric and heteromeric protein complexes are ubiquitous across all domains of life. The evolutionary transition from homo- to hetero-oligomers by gene duplication and chain specialization is widespread, yet it entails challenging requirements for maintaining oligomerization and functionality. Chain specialization in ferritins, which occurs in bacteria, vertebrates, and plants, is a salient example of this phenomenon. In heteroferritins, the two essential functions, ferroxidase activity and electron transfer, are split between two specialized chain types. Many heteroferritins assemble into complexes with variable subunit ratios, implying the existence of assembly rules that balance compositional flexibility with structural constraints. Here, we identify the assembly rules governing the organization of the heterobacterioferritin from Magnetospirillum gryphiswaldense (MSR-1 Bfr) by analyzing its cryo-EM reconstructions. These rules consist of structural constraints that limit the number of possible arrangements and promote juxtaposition of the two, now separated functions. These constraints support compositional flexibility while preserving function, thereby providing resilience to stochastic variation in oligomer stoichiometry. Bioinformatic analysis revealed that the assembly rules identified in MSR-1 Bfr are widespread across the Bfr family and coevolved with chain specialization. Together, these findings support leading models of hetero-oligomer evolution and reveal the emergence of order-exerting mutations that shape the organization of multimeric protein complexes while conserving function.
|

|
Scooped by
mhryu@live.com
Today, 5:05 PM
|
β-Elemene, a sesquiterpene with anticancer activity, faces limited microbial production due to low yields and inefficient enzyme coordination. This study established a modular covalent enzyme cascade in E. coli for high-efficiency β-elemene biosynthesis. Systematic screening identified the SnoopTag/SnoopCatcher-mediated covalent assembly of farnesyl diphosphate synthase and germacrene A synthase as the optimal configuration (strain FS07). Subsequent enzyme stoichiometry modulation or linker engineering did not surpass FS07’s performance, indicating a near-optimal design. Fermentation optimization elevated the β-elemene titer to 7.21 g/L in FS07. Fed-batch fermentation in a 1.3 L bioreactor subsequently increased the final titer to 31.21 g/L, representing a 4-fold improvement over scaffold-free controls and achieving the highest reported yield in E. coli to date. This work provided a robust enzymatic scaffolding strategy for high-level terpenoid production.

|
Scooped by
mhryu@live.com
Today, 4:57 PM
|
Glucose is an important substrate for organisms to acquire energy needed for cellular growth. Despite the importance of this metabolite, single-cell information at a fast time-scale about the dynamics of intracellular glucose levels is difficult to obtain as the current available sensors have drawbacks in terms of pH sensitivity or unmatched glucose affinity. To address this, we developed a convenient method to create and screen biosensor libraries using yeast as a workhorse. This resulted in TINGL (Turquoise INdicator for GLucose), a robust and specific biosensor with an affinity that is compatible with intracellular glucose detection. We show that the sensor can be calibrated in vivo (i.e., intracellular) through equilibration of internal and external glucose in a yeast mutant unable to phosphorylate glucose. Using this method, we estimated dynamic glucose levels in budding yeast during transitions to glucose. We found that glucose concentrations reached levels up to approximately 1 mM as previously determined biochemically. Furthermore, the sensor showed that intracellular glucose dynamics differ based on whether cells are glucose-repressed or not. Finally, the human codon-optimized version (THINGL, Turquoise Human INdicator for GLucose) also showed a robust response after glucose addition to starved human cells, showing the versatility of the sensors. We believe that this sensor can aid researchers interested in cellular carbohydrate metabolism.

|
Scooped by
mhryu@live.com
Today, 4:37 PM
|
Probiotics exert many effects through probiotic effector molecules (PEMs), which are secreted or surface-associated bioactive compounds. Key classes of PEMs include bacterial glycan polymers (e.g., exopolysaccharides), surface proteins and pili, secreted peptides and enzymes, extracellular vesicles, and small-molecule metabolites. These bioactive compounds mediate host–microbe crosstalk, reinforcing epithelial barrier integrity, shaping gut microbial communities, and modulating immune responses. Their production is strain-specific and influenced by environmental conditions, whereas their activities depend on receptor interactions such as with Toll-like receptors, G protein–coupled receptors, and aryl hydrocarbon receptors. Major challenges include high-throughput identification of novel PEMs, in situ verification of their gut production, and determination of effective doses. Emerging approaches, including comparative genomics, synthetic biology, and next-generation probiotics, promise to unlock PEMs’ therapeutic potential. A mechanistic understanding of PEM diversity and function will facilitate the design of targeted probiotic therapies and innovative functional foods.

|
Scooped by
mhryu@live.com
Today, 4:07 PM
|
Inorganic phosphate (Pi) is essential for all living organisms. PstSCAB, a bacterial high-affinity ABC transporter, imports Pi under limiting conditions via five subunits: PstA and PstC forming the transmembrane domain (TMD), periplasmic PstS that switches between free and TMD-docked forms for Pi capture and delivery, and two cytosolic PstB subunits for ATP binding and hydrolysis. Its malfunction affects the virulence of pathogenic bacteria, making it pharmaceutically attractive. However, complete structural pictures of PstSCAB in different states remain lacking. Here, we determine cryo-EM structures of PstSCAB in resting, pretranslocation, and catalytic intermediate states, which reveal that conformational changes in PstS and ATP binding/unbinding in PstB collectively induce rigid-body movements of TMD, generating inward- or outward-facing conformations. In TMD, Pi specificity is determined by positively charged Arg220 (PstA) and Arg237 (PstC). This study advances understanding of bacterial Pi import and supports drug development targeting PstSCAB. Bacterial phosphate transporter PstSCAB is essential for survival and virulence. Here, authors reveal cryo-EM structures of PstSCAB in multiple states, uncovering how conformational changes drive phosphate import and providing insights for antibiotic development.

|
Scooped by
mhryu@live.com
Today, 3:59 PM
|
Neuronal networks have driven advances in artificial intelligence, while molecular networks can provide powerful frameworks for energy-efficient information processing. Inspired by biological principles, we present a computational framework for mapping synthetic gene circuits into bio-inspired electronic architectures. In particular, we developed logarithmic Analog-to-Digital Converter (ADC), operating in current mode with a logarithmic encoding scheme, compresses an 80 dB dynamic range into three bits while consuming less than 1 µW, occupying only 0.02 mm², and operating at 4 kHz. Our bio-inspired approach achieves linear scaling of power, unlike conventional linear ADCs where power consumption increases exponentially with bit resolution, significantly improving efficiency in resource-constrained settings. Through a computational trade-off analysis, we demonstrate that logarithmic encoding maximizes spatial resource efficiency among power consumption and computational accuracy. By leveraging synthetic gene circuits as a model for efficient computation, this study provides a platform for the convergence of synthetic biology and bio-inspired electronic design. Ilan Oren and colleagues present a synergistic framework that translates gene circuits into energy-efficient electronic circuits, with a focus on data converters. Trade-off analysis reveals that logarithmic encoding optimizes spatial efficiency among power consumption and computational accuracy.

|
Scooped by
mhryu@live.com
Today, 3:39 PM
|
Bacterial cellulose possesses excellent biocompatibility and mechanical strength but lacks the bioactivity needed for many biomedical and healthcare applications. To address this limitation, we develop a metabolic glycoengineering–click chemistry strategy that enables in situ incorporation of azide groups into bacterial cellulose, followed by mild and selective conjugation of alkyne-bearing functional molecules. This approach avoids harsh chemical treatments, preserves the native properties of bacterial cellulose, and supports stable attachment of diverse bioactive agents, including antibacterial porphyrins, arginine-glycine-aspartic acid peptides, and recombinant proteins with fluorescent or enzymatic functions. As a proof-of-concept, a cascade catalytic system comprising glucose oxidase and superoxide dismutase is immobilized onto azide-modified bacterial cellulose, yielding a multifunctional wound dressing designed to address hyperglycemia and oxidative stress—key barriers to chronic wound healing. In male diabetic mice, this glucose oxidase/superoxide dismutase-integrated bacterial cellulose dressing (low endotoxin <0.1 EU/mL) accelerates wound closure to 92.1% by day 14, significantly outperforming the controls. Our strategy highlights a scalable and bio-orthogonal route for enhancing bacterial cellulose with user-defined bioactivities, thereby expanding its utility in advanced biomaterials development. Bacterial cellulose (BC) possesses excellent biocompatibility and mechanical strength but lacks the bioactivity needed for many biomedical applications. Here the authors use glycoengineering–click chemistry to incorporate azide groups into BC in situ, and show enhanced BC-mediated wound healing.

|
Scooped by
mhryu@live.com
Today, 3:17 PM
|
Enterohemorrhagic E. coli (EHEC) is a severe foodborne pathogen that can lead to hemolytic uremic syndrome. However, antibiotics are contraindicated for EHEC treatment due to toxin release and gut microbiota disruption. Here we report a dual‑mechanism therapeutic strategy combining an engineered E. coli Nissle 1917 strain (EcN3) with 2′‑fucosyllactose (2‑FL) delivered via multicompartment microspheres (MCMs). EcN3 expresses α‑L‑fucosidase to hydrolyze 2‑FL into lactose and fucose. Lactose enhances glucuronic acid utilization, limiting a preferred nutrient of EHEC, whereas fucose activates FusKR signaling to suppress virulence gene expression. MCMs confer gastric protection and enable targeted colonic release, ensuring coordinated activity. In female mouse models and infant rabbit models of Citrobacter rodentium and EHEC infection, this system reduces intestinal colonization, virulence gene expression and epithelial damage without inducing Shiga toxin production. Moreover, MCMs-based strategy preserves the relative abundance of Lactobacillus, and promotes intestinal integrity. This targeted strategy presents a viable alternative to antibiotics, addressing EHEC pathogenesis and antibiotic resistance. Enterohemorrhagic EHEC is a foodborne pathogen where therapeutic antibiotic treatment can release toxins and disrupt gut microbiota. Here the authors demonstrate a therapeutic strategy where an engineered Escherichia coli Nissle 1917 limits the glucuronic acid utilization by EHEC.

|
Scooped by
mhryu@live.com
Today, 11:42 AM
|
Regulating gene expression with precision is essential for cellular engineering and biosensing applications, where rapid, programmable, and sensitive control is desired. Current approaches to regulatory circuit design often rely on control at a single regulatory level, primarily the transcriptional level, thereby limiting the capability of fine-tuning the regulatory dynamics in response to complex stimuli. To address this challenge, we developed four novel RNA-protein hybrid type-1 incoherent feed-forward loop (I1-FFL) circuits in E. coli that integrate transcriptional and translational regulators to achieve multilevel control of gene expression. These hybrid circuits leverage the modularity and rapid dynamics of RNA-based activators alongside the versatile inhibition capabilities of the protein-based repressors to endow tunable pulse dynamics through engineered delays that act as transient repressor decoys. By repurposing synthetic RNA regulators at multiple regulatory levels together with aptamers and RNA-binding proteins, we demonstrate previously unexplored circuits with tunable dynamics. Complementary simulation results highlighted the importance of the engineered delays in achieving tunable pulse dynamics in these circuits. Integrating modeling insights with experimental validation, we demonstrated the flexibility of designing the RNA-protein hybrid I1-FFL circuits, as well as the tunability of their dynamics, highlighting their suitability for applications in environmental monitoring, metabolic engineering, and other engineered biological systems where precise temporal control and adaptable gene regulation are desired.

|
Scooped by
mhryu@live.com
Today, 11:27 AM
|
PAR-CLIP is a widely used method for identifying binding sites of RNA-binding proteins (RBPs) transcriptome-wide. A characteristic T-to-C transition in the sequenced complementary DNA pinpoints the site of RBP-RNA crosslinking and is induced by the use of a photoreactive uridine analogue, 4-thiouridine (4SU). As with other system-wide methods, PAR-CLIP, too, is prone to false discoveries, as the T-to-C signal might result from systematic noise, pre-existing SNPs, and polymerase chain reaction errors. Therefore, rigorous statistical methods are required for analyzing PAR-CLIP data. The few existing tools to analyze PAR-CLIP data lack updates and sufficient documentation, and often fail to process current higher-depth sequencing data. Here, we report PCLIPtools, a lightweight, customizable suite for analyzing PAR-CLIP data. PCLIPtools considers the read depth, T-to-C transitions, and the other mutations to statistically estimate high-confidence interaction sites. Benchmarking shows that PCLIPtools identifies more functionally significant targets than the current standard tool, PARalyzer, without losing high-confidence sites and outperforming it in runtime. Exploratory analyses show PCLIPtools’ specific targets are enriched for read depth and T-to-C conversion, supporting their validity. With simplicity, robustness, and speed, PCLIPtools improves the precision of PAR-CLIP data analysis while being accessible to experimental RNA biologists.

|
Scooped by
mhryu@live.com
Today, 9:42 AM
|
Protein aggregation poses a significant risk to biopharmaceutical product quality, as even minor amounts of oligomeric species can compromise efficacy and safety. Rapid and reliable detection of protein aggregates thus remains a major challenge in biopharmaceutical manufacturing. Although traditional offline methods such as size-exclusion chromatography provide accurate results, their inherent time delays limit real-time process control capabilities. Consequently, there is an urgent scientific need for inline analytical techniques capable of selectively quantifying protein monomers and aggregates in real time to facilitate immediate corrective actions and enhance overall process robustness. Raman spectroscopy, as a tool for a process analytical technology application, is especially suitable due to its molecular specificity, rapid data acquisition, and compatibility with aqueous solutions commonly used in biopharmaceutical manufacturing. Addressing this need, this study establishes a Raman spectroscopy-based strategy for the selective detection and quantification of monomeric and aggregated forms of a model protein (bovine serum albumin). Controlled stress conditions were applied to generate aggregated species reproducibly, and a Latin Hypercube sampling design was used to independently vary protein concentration and aggregate fraction, ensuring that observed spectral effects were attributable to aggregation rather than concentration differences. Furthermore, spectral markers identified in spectra acquired from multiple chromatographic runs were qualitatively compared with offline reference measurements from size-exclusion chromatography. This limitation in real-time applicability was circumvented by chemometric machine learning approaches. The use of convolutional neural networks enabled the selective quantification of the protein monomers and aggregates and delivered superior predictive performance and robustness across cross-validation, independent testing, and synthetic perturbation scenarios compared to traditional chemometric approaches. Collectively, these results demonstrate that the selected Raman spectral markers, combined with advanced chemometric modeling, enable reliable, real-time monitoring of protein size variants in biopharmaceutical downstream processes.

|
Scooped by
mhryu@live.com
Today, 1:56 AM
|
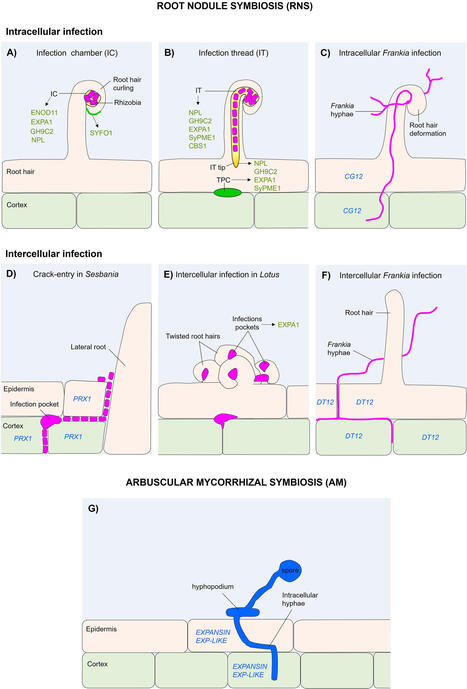
Plant roots are usually ground organs that perform essential roles, mostly associated with the anchoring of plants to the soil and absorption of nutrients and water. However, they are also exposed to a wide variety of microorganisms and may develop various symbiotic relationships, such as mutualism, which benefits both organisms. For instance, arbuscular mycorrhizal AMF symbiosis is likely the oldest and most widespread mutualistic association, that occurs between plants and fungi. Another relevant example is the root nodule symbiosis, established between nitrogen-fixing bacteria and nodulating legumes, actinorhizal plants and Parasponia species. In both cases, microbial colonization of plant roots culminates in the formation of specialized symbiotic structures. In this regard, microbial infection is a critical step for the mutualistic relationship, where altering the cell wall biomechanics is necessary to facilitate microbial entry, which can be modulated by various cell wall protein families. This review examines the current knowledge on cell wall modifications occurring in plants roots during the symbiotic entry of microorganisms, focusing on the role of cell wall-remodeling proteins involved in these processes.

|
Scooped by
mhryu@live.com
Today, 1:41 AM
|
Cultured meat, produced through in vitro cultivation of animal cells, represents a promising solution to the environmental, ethical, and resource-intensive challenges of conventional livestock production. Significant technological advances have been achieved in seed cell acquisition, serum-free culture media development, bioreactor scale-up, 3D tissue formation, and sensory enhancement. Nevertheless, substantial hurdles persist in achieving cost-effective large-scale production, ensuring product safety and quality, establishing robust regulatory frameworks, and securing widespread consumer acceptance. This review examines recent progress throughout the cultured meat production chain, critically analyzes unresolved challenges, and outlines essential research priorities for realizing its industrial potential.
|
 Your new post is loading...
Your new post is loading...



































ALG-GOX bulk hydrogel synthesis