Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:58 AM
|
There is still a need for a better understanding of how abiotic/biotic factors affect the functional structure and composition of biological assemblages, given that living organisms are in constant interaction with their environment and with each other. Here, we present a comprehensive dataset of 31 functional traits of bacteria using information from BacDive, a bacterial diversity meta-database, as well as from the rrnDB and genomesizeR datasets. This updated version of the BactoTraits dataset, in addition to now offering more traits for more strains (97,721 strains with at least one trait described), makes R scripts available to the scientific community. These traits include physiological characteristics, metabolic processes, genome properties and biotope preferences. They could be inferred to the whole bacterial community thanks to taxonomic affiliation obtained from traditional high throughput 16S rRNA gene amplicon sequencing methods. This taxonomic affiliation is based on the regularly updated SILVA database and thus allows to study combinations of weighted mean trait profiles of bacterial communities at different taxonomic levels. BactoTraits can be used, for example, to improve predictions of ecological responses to natural/anthropogenic pressures and to support biomonitoring, management and conservation strategies. The R scripts, as well as the dataset encoded in BactoTraits, are available at: https://doi.org/10.24396/ORDAR-182.

|
Scooped by
mhryu@live.com
Today, 1:45 AM
|
Plants and microbes exchange macromolecules such as RNA and proteins. How this exchange is accomplished is poorly understood, but extracellular vesicles (EVs) have been proposed as likely vehicles. Here, we review recent work on the biogenesis and functions of plant EVs and the current evidence in support of and against their role in cross-kingdom RNA interference. Plant EVs, like EVs from other kingdoms of life, are released in part by the fusion of multivesicular bodies with the plasma membrane, a complex and conserved mechanism involving lipid-modifying proteins, the exocyst complex, and Rab GTPases. Though some plant EV subpopulations are involved in immunity, it appears unlikely that plant EVs contribute to cross-kingdom RNA interference. Recent work has shown that plants secrete extravesicular RNA, including small RNAs and long noncoding RNAs, into the leaf apoplast and onto leaf surfaces, while very little RNA is found inside of EVs. We propose that these free extracellular RNAs play a central role in maintaining a healthy leaf microbiome.

|
Scooped by
mhryu@live.com
Today, 1:29 AM
|
Homomeric and heteromeric protein complexes are ubiquitous across all domains of life. The evolutionary transition from homo- to hetero-oligomers by gene duplication and chain specialization is widespread, yet it entails challenging requirements for maintaining oligomerization and functionality. Chain specialization in ferritins, which occurs in bacteria, vertebrates, and plants, is a salient example of this phenomenon. In heteroferritins, the two essential functions, ferroxidase activity and electron transfer, are split between two specialized chain types. Many heteroferritins assemble into complexes with variable subunit ratios, implying the existence of assembly rules that balance compositional flexibility with structural constraints. Here, we identify the assembly rules governing the organization of the heterobacterioferritin from Magnetospirillum gryphiswaldense (MSR-1 Bfr) by analyzing its cryo-EM reconstructions. These rules consist of structural constraints that limit the number of possible arrangements and promote juxtaposition of the two, now separated functions. These constraints support compositional flexibility while preserving function, thereby providing resilience to stochastic variation in oligomer stoichiometry. Bioinformatic analysis revealed that the assembly rules identified in MSR-1 Bfr are widespread across the Bfr family and coevolved with chain specialization. Together, these findings support leading models of hetero-oligomer evolution and reveal the emergence of order-exerting mutations that shape the organization of multimeric protein complexes while conserving function.

|
Scooped by
mhryu@live.com
Today, 1:06 AM
|
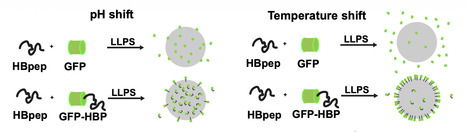
Cells contain membrane-bound and membraneless organelles that operate as spatially distinct biochemical niches. However, these reaction centers lose fidelity due to aging or diseases. A grand challenge for biomedicine is restoring or augmenting cellular functionalities. An excited strategy is the delivery of protein-based materials that can directly interact with cellular biological networks. In this study, we sought to develop long-lasting materials capable of cellular uptake, akin to intracellular interaction hubs. We develop a delivery method to efficiently transplant stable micron-size peptide-based compartments into living cells. By loading coacervates with nanobodies and bioPROTACs, we demonstrate successful target sequestration of natively expressed GFP to our synthetic hubs, and function as bioreactors to selectively degrade GFP inside human cells. These results represent an important step toward the development of synthetic organelles that can be fabricated in vitro and taken up by cells for applications in cell engineering and regenerative medicine. Short HBpep peptides assemble into micron-size coacervates that are efficiently taken up by various cell types and stably retained for days. Nanobodies and bioPROTACs loaded in the coacervates enable interaction with native targets and these hubs can function as a bioreactor for target degradation

|
Scooped by
mhryu@live.com
Today, 12:48 AM
|
Proteases contribute to essential cellular processes through catalyzing proteolysis, resulting in peptide bond hydrolysis and the generation of novel polypeptide species. Identification of proteolytic cleavage events is crucial for discerning proteolytic networks in biological systems, including the contribution of individual proteases to specific disease states. As such, various mass spectrometry-based workflows have been exploited for the sensitive identification of cleavage sites. To date, a range of enrichment strategies have been developed, focusing on increasing sensitivity, ease, and throughput for protease substrate discovery. Recent advances in mass spectrometry instrumentation have also permitted enrichment-free workflows for degradomics analysis, providing simultaneous and systematic assessment of the proteome and degradome. In this review, we discuss current technologies for the enrichment and identification of both N- and C-termini, as well as their application to profile protease specificity and decipher individual substrate repertoires in diverse biological conditions.

|
Scooped by
mhryu@live.com
Today, 12:40 AM
|
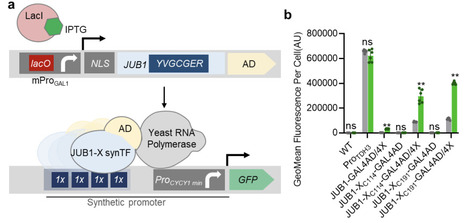
Technologies developed over the past decade have made Saccharomyces cerevisiae a promising platform for producing various natural products. Balancing multi-enzyme expression, while maintaining robust microbial growth, remains a limiting factor for engineering long biosynthetic pathways in yeast. Here, we improved the transcriptional capacity of our previously developed IPTG-inducible synthetic transcription factors (synTFs) derived from the plant JUB1 DNA-binding domain. To this end, at cysteine positions within surface-exposed loop regions of a JUB1-derived DNA-binding scaffold, we introduced a short peptide to enhance loop flexibility while providing local stability and orientation. The generated synTFs, so-called JUB1-X synTFs, varying in strength, have been successfully used to improve the synthesis of 3′-phosphoadenosine 5′-phosphosulfate (PAPS), a universal sulfate donor necessary for the synthesis of bioactive molecules, including therapeutic glycosaminoglycans and sulfolipids. Using only this engineered yeast strain in simple batch culture, PAPS accumulation of 21.4 ± 5.8 mg/g cdw was achieved after only 5 hours of inducing the expression of JUB1-X synTFs. Beyond PAPS production, the design principle demonstrated here provides a generalizable strategy to fine-tune other plant-derived synTFs, expanding the regulatory capabilities of existing synTF collections. Together, this work offers a modular, scalable approach to constructing high-performance gene circuits and supports the development of yeast cell factories for complex metabolic and synthetic biology applications.

|
Scooped by
mhryu@live.com
Today, 12:24 AM
|
Despite the fact that microbes in natural environments spend most of their time in growth arrest, we understand little about how this physiological state affects the performance of engineered genetic circuits. Here, we measure repression curves from a library of genetic NOT gates at single-cell resolution in Escherichia coli under both active growth and growth arrest to systematically investigate how growth arrest affects circuit behavior. We find that the impact of growth arrest on circuit performance is almost entirely dominated by a single effect: a >100-fold reduction in unrepressed expression levels. Growth arrest caused gene expression noise to increase moderately and had only minimal impacts on the sensitivity and sharpness of the repression curves. Our work shows both that conventional genetic circuit design paradigms are currently insufficient to develop circuits that can function properly under growth arrest, but also that addressing the reduction in just a single performance parameter would be sufficient to resolve this problem. This work expands our understanding of bacterial gene regulation under growth arrest and lays the groundwork for new design paradigms that will be essential in ensuring the safe and reliable performance of synthetic biology systems in real-world environments.

|
Scooped by
mhryu@live.com
February 2, 11:47 PM
|
Energy homeostasis facilitated by the interplay of substrate-level and oxidative phosphorylation is crucial for bacterial adaptation to diverse substrates and environments. To investigate how bioenergetic systems optimize under metabolically restrictive conditions, we evolved electron transport system (ETS) variants with distinct proton-pumping efficiencies on succinate and glycerol. These substrates impose unique metabolic constraints: succinate requires complete gluconeogenesis, while glycerol supports mixed glycolytic and gluconeogenic fluxes. Multi-scale computational analysis of the strains revealed (a) Growth optimization across carbon substrates for multiple ETS variants, (b) A conserved aerobicity stimulon comprising seven independently regulated gene groups that are co-regulated with increasing aerobic capacities, (c) Proteome reallocation linked to aerobicity, validated using genome-scale metabolism and expression modeling, and (d) Carbon source-specific compensatory mutations. These findings define the aerobicity stimulon and establish a unifying framework for understanding bacterial respiratory flexibility, demonstrating how transcriptional networks and metabolic systems integrate to achieve energy homeostasis and bioenergetic resilience.

|
Scooped by
mhryu@live.com
February 2, 11:34 PM
|
The gut microbiota plays a crucial role in insect immune priming, inducing enhanced immune response that functionally resembles acquired immunity confined to vertebrates. While gut microbiota mediates systemic immune activation in insect hemolymph, the mechanisms underlying remote immunoregulation remain largely unknown. Here we use the honey bee gut microbiota as a model, we identify butyrate as a key microbial metabolite coordinating immune-metabolic crosstalk. Butyrate supplementation restores immune competence in germ-free bees, mirroring the protective effects of microbiota-colonized individuals. Butyrate orchestrates lipid metabolic reprogramming in the fat body by activating glycerolipid and arachidonic acid metabolism through activating the G-protein coupled receptor 41 while inhibiting histone deacetylases. These changes in-turn upregulate prostaglandin E2 biosynthesis, which is essential for humoral and cellular immune activation. These findings unravel how the intricate integration of immune and metabolic systems in honey bees is driven by gut-host interactions. The intestinal microbiota is shown to play a critical role in immune priming in insects. Here the authors show that microbiota-derived butyrate is linked to the priming of systemic immune responses via lipid metabolic reprogramming in honey bees.

|
Scooped by
mhryu@live.com
February 2, 7:26 PM
|
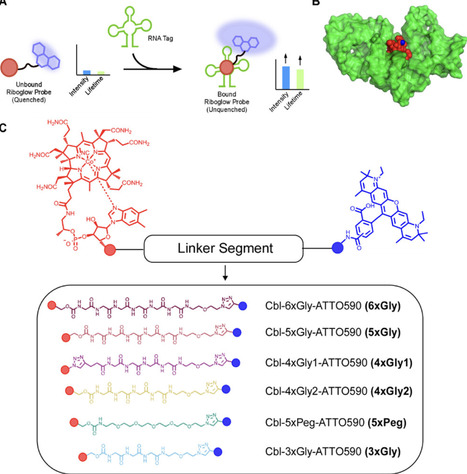
Riboglow probes are small molecules where a synthetic fluorophore is connected to an RNA-binding moiety via a chemical linker. Upon binding a short RNA sequence, probe fluorescence intensity and lifetime increase. The fluorescence change is modulated by the architecture of the chemical linker. Here, we systematically interrogated the linker composition in a series of Riboglow probes and assessed fluorescence properties. We found that glycine linkers result in higher fluorescence turn-on compared to a polyethylene glycol linker of similar length. When varying the length of the polyglycine linker, we found that increasing the number of glycine residues led to more substantial fluorescence turn-on upon RNA-ligand binding. Surprisingly, the composition of the Riboglow chemical linker influences fluorescence lifetime contrast when comparing probe binding to two different RNA ligands, a quality necessary for RNA multiplexing. Finally, evaluating probe fluorescence lifetimes in live mammalian cells demonstrated the ability of new Riboglow probes to visualize RNAs live. Insights gained from the systematic assessment of the linker’s architecture will dictate the rational design of future fluorophore-quencher probe designs.

|
Scooped by
mhryu@live.com
February 2, 4:58 PM
|
Antibiotic resistance (AR) is an escalating public health threat, necessitating innovative strategies to control resistant bacterial populations. One promising approach involves engineering genetic elements that can spread within microbial communities to eliminate AR genes. Previously, we developed Pro-Active Genetics (Pro-AG), a CRISPR-based gene-drive-like system capable of reducing AR colony-forming units (CFU) by approximately five logs. Here, we advance this technology by integrating Pro-AG into a conjugative transfer system, enabling efficient dissemination of an anti-AR gene cassette between two bacterial strains. Additionally, we characterize a complementary homology-based deletion (HBD) process, a CRISPR-driven mechanism that precisely removes target DNA sequences flanked by short direct repeats. Our findings reveal that Pro-AG and HBD are differentially influenced by the bacterial RecA pathway and that HBD components can be delivered via plasmids or phages to selectively delete Pro-AG cassettes. This built-in safeguard prevents uncontrolled spread of a gene cassette and mitigates unanticipated side effects. These refinements enhance the efficiency and flexibility of Pro-AG, expanding its potential applications in microbiome engineering, environmental remediation, and clinical interventions aimed at combating antibiotic resistance. More broadly, this work establishes a proof-of-principle for microbiome engineering strategies that could be leveraged to improve health and restore ecological balance.

|
Scooped by
mhryu@live.com
February 2, 4:46 PM
|
The increasing imperative to mitigate greenhouse gas emissions and foster the transition to a low-carbon bioeconomy has intensified interest in methane bioconversion as a sustainable approach for transforming methane into valuable bioproduction. Although advancements have been made in optimizing methanotrophic pathways to improve bioproduction, significant challenges persist, including methane solubility, bioavailability, and metabolic flexibility, limiting the efficiency of methane conversion. This review provides a comprehensive overview of the initiatives aimed at developing next-generation methanotrophic cell factories by overcoming the physiological limitations of natural methanotrophs. We first analyze the metabolic characteristics of methanotrophs for assimilating methane into cellular building blocks. Then, we discuss methane assimilation pathways and their unique characteristics in matter and energy transmission for facilitating the integration of methane into central carbon metabolism. Further, we propose a systematic framework for designing methane-based biomanufacturing to enable low-carbon bioproduction by integrating synthetic biology, metabolic engineering, and systems biology, thereby developing efficient methane assimilation cell factories for producing high-value bioproducts. Finally, we prospect the potential for valorizing methane derived from anthropogenic emissions and renewable sources, while identifying the key challenges and future research directions necessary for advancing a sustainable, low-carbon bioeconomy.

|
Scooped by
mhryu@live.com
February 2, 4:31 PM
|
Antibiotic resistance is escalating, highlighting the urgent need for novel antimicrobial strategies. Defensin-like antimicrobial peptides (AMPs) are considered ideal candidates due to their broad-spectrum activity and engineerable potential; however, their limited antimicrobial efficacy and complex chemical synthesis constrain practical applications. In this study, we aimed to enhance the antimicrobial properties of defensin-like AMP through rational design, directed evolution, and structural fusion strategies. The engineered variant XC1 demonstrated significantly improved antimicrobial activity against a broad range of pathogens, including methicillin-resistant Staphylococcus aureus, while maintaining broad-spectrum efficacy. Comprehensive evaluation of toxicity and stability showed that XC1 exhibited good functional stability in serum, low hemolysis, and low cytotoxicity, indicating excellent therapeutic potential. In addition, high-level secretory expression of defensin-derived AMPs and their engineered variants was achieved using Pichia pastoris GS115, demonstrating strong biosynthetic capability. Together, these results provide a viable strategy for enhancing the antimicrobial activity and scalable biosynthesis of defensin-like AMPs.
|

|
Scooped by
mhryu@live.com
Today, 1:56 AM
|
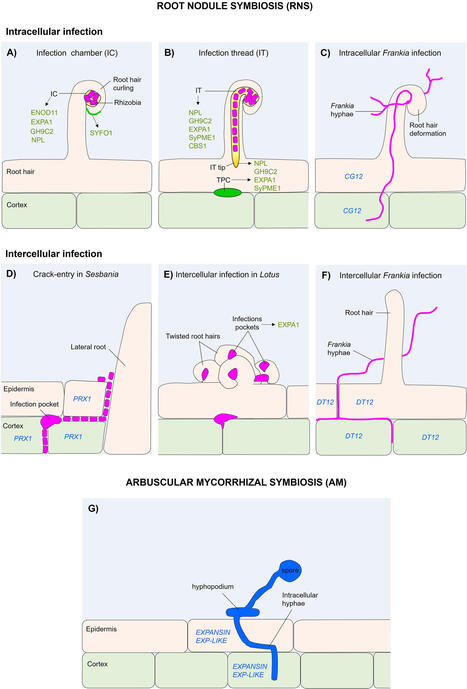
Plant roots are usually ground organs that perform essential roles, mostly associated with the anchoring of plants to the soil and absorption of nutrients and water. However, they are also exposed to a wide variety of microorganisms and may develop various symbiotic relationships, such as mutualism, which benefits both organisms. For instance, arbuscular mycorrhizal AMF symbiosis is likely the oldest and most widespread mutualistic association, that occurs between plants and fungi. Another relevant example is the root nodule symbiosis, established between nitrogen-fixing bacteria and nodulating legumes, actinorhizal plants and Parasponia species. In both cases, microbial colonization of plant roots culminates in the formation of specialized symbiotic structures. In this regard, microbial infection is a critical step for the mutualistic relationship, where altering the cell wall biomechanics is necessary to facilitate microbial entry, which can be modulated by various cell wall protein families. This review examines the current knowledge on cell wall modifications occurring in plants roots during the symbiotic entry of microorganisms, focusing on the role of cell wall-remodeling proteins involved in these processes.

|
Scooped by
mhryu@live.com
Today, 1:41 AM
|
Cultured meat, produced through in vitro cultivation of animal cells, represents a promising solution to the environmental, ethical, and resource-intensive challenges of conventional livestock production. Significant technological advances have been achieved in seed cell acquisition, serum-free culture media development, bioreactor scale-up, 3D tissue formation, and sensory enhancement. Nevertheless, substantial hurdles persist in achieving cost-effective large-scale production, ensuring product safety and quality, establishing robust regulatory frameworks, and securing widespread consumer acceptance. This review examines recent progress throughout the cultured meat production chain, critically analyzes unresolved challenges, and outlines essential research priorities for realizing its industrial potential.

|
Scooped by
mhryu@live.com
Today, 1:19 AM
|
Lung cancer is the leading cause of cancer-related mortality worldwide. It is frequently diagnosed at an advanced stage and exhibits significant morphological and molecular heterogeneity, encompassing both small- and non-small-cell types, as well as major histological variations. Conventional imaging and tissue biopsies are invasive and may fail to reflect dynamic tumor biology. Exosomes (30-150 nm, extracellular vesicles that are products of the endosome) are present in plasma, serum, saliva, and urine and contain tumor-reflective proteins, lipids, and RNAs. This review summarizes the strategies used in biosensors to detect exosomes in lung cancer using various approaches, including electrochemical, optical, microfluidic, and nanomaterial-assisted biosensors, which detect various markers, such as PD-L1, EGFR, and oncogenic miRNAs/lncRNAs. Exosomes possess inherent biocompatibility, intrinsic cargo protection, and the ability to target tumors (homing capabilities), attributes that synthetic nanocarriers, such as liposomes, polymeric, and inorganic nanoparticles, lack. These characteristics render exosomes advantageous for biosensing and delivery applications. We also discuss exosome-enabled therapeutic directions, such as drug- and nucleic-acid-loaded exosomes and immunomodulatory cargos, and indicate the new clinical-study environment. The most important obstacles are the non-uniformity of vesicles, variability in isolation/quantification, and lack of standardization. The discussion concludes with an examination of the practice performance standards, reporting priorities, and emerging trends. These include AI-assisted analysis and the use of portable point-of-care devices, which aim to expedite the development of clinically deployable exosome platforms for lung cancer diagnosis. We examined the integration of a sample into an answer, validation of assays on cohorts, and conceptual implications of regulatory considerations for reproducible measurements. This approach facilitates practical applications, such as screening, treatment monitoring, and early relapse detection.

|
Scooped by
mhryu@live.com
Today, 12:54 AM
|
Circular mRNA faces challenges in enhancing its translation potential as an RNA therapeutic. Here we introduce two molecular designs that bolster circular mRNA translation through an internal cap-initiated mechanism. The first consists of a circular mRNA with a covalently attached N7-methylguanosine (m7G) cap through a branching structure (cap-circ mRNA). This modification allows circular mRNA to recruit translation machinery and produce proteins more efficiently than internal ribosome entry site (IRES)-containing circular mRNAs. Combining with an N1-methylpseudouridine (m1Ψ) modification, cap-circ mRNA exhibits a lower acute immunostimulatory effect, maintaining high translation in mice. The second design features the non-covalent attachment of an m7G cap to a circular mRNA through hybridization with an m7G cap-containing oligonucleotide, enhancing translation by more than 50-fold. This setup allows circular mRNAs to synthesize reporter proteins upon hybridizing with capped mRNAs or long non-coding RNAs and to undergo rolling circle-type translation. These advancements broaden the therapeutic applications of circular mRNAs by minimizing their molecular size, elevating translation efficiency and facilitating cell-type-selective translation. Placement of an m7G cap internally on circular RNAs promotes their translation in vivo.

|
Scooped by
mhryu@live.com
Today, 12:45 AM
|
Diverse bacterial sensory domains detect intracellular and extracellular cues and relay this information to downstream signaling pathways. PAS domains are well known to bind cofactors such as heme for intracellular sensing, whereas Cache domains, the largest family of bacterial extracytoplasmic sensor modules, are thought to function exclusively as cofactor independent ligand binding domains. Here, we identify and characterize a large family of Cache domains that contain a covalently bound c type heme. Systematic sequence analysis of the DUF3365/Tll0287 family of unknown function reveals exceptional conservation of a canonical CXXCH heme-binding motif, establishing c-type heme binding as a defining feature of this domain family. Structure- and sequence-based comparisons demonstrate that, despite prior annotation as PAS, these domains belong to the Cache superfamily and share a conserved Cache fold augmented by a distinct structural element harboring the heme-binding motif. Heme containing Cache domains are widely distributed across bacterial phyla, with strong enrichment in Gracilicutes, and frequently occur as stand-alone modules, an atypical presentation of Cache domains. Phylogenetic analyses support insertion of a short cytochrome c derived fragment containing the heme-binding motif into a preexisting Cache scaffold, representing a previously unrecognized mode of sensor domain evolution. Spectral and ligand binding analyses indicate that heme containing Cache domains fused to signal-transduction modules have properties typical of nitric oxide, but not oxygen, sensors; whereas stand-alone domains likely have redox-related roles. Our findings expand the functional repertoire of Cache domains to include covalently bound cofactors and reveal fragment-level recruitment of enzymatic motifs as a route for the emergence of bacterial sensory systems.

|
Scooped by
mhryu@live.com
Today, 12:28 AM
|
Single-cell phenotypic diversity underlies fundamental bacterial behaviors. Resolving this heterogeneity at scale while maintaining sensitive detection remains a central challenge. Here, we address this gap by combining the strengths of single-molecule fluorescence in situ hybridization (smFISH) and transcriptome-wide sequencing. We present par2FISH, a framework for discovering transcriptionally defined subpopulations across conditions by multiplexing tens of thousands of smFISH reactions. Through comparative single-cell transcriptomics in E. coli and Salmonella, we uncover conserved and species-specific patterns of heterogeneity, including mutually exclusive states, coordinated metabolic specialization, and differential activation of virulence programs. We further obtain complete transcriptomes for selected subpopulations using marker-based cell sorting and RNA-sequencing (FACS-RNA-seq). This integrated strategy paves the way for exposing bacterial phenotypic landscapes and the environmental, physiological, and evolutionary factors that shape them.

|
Scooped by
mhryu@live.com
Today, 12:09 AM
|
Dopamine deficiency resulting from nigrostriatal dopaminergic neuronal damage manifests as extrapyramidal motor symptoms of Parkinson’s disease (PD). Oral tablet dosing of levodopa, administered 3–4 times a day, remains the standard of care due to its tolerability and effectiveness; however, it is prone to deleterious side effects, including off-periods and levodopa-induced dyskinesia after long-term use. Herein, using synthetic biology approaches, we developed and systematically evaluated the feasibility of a probiotic-based live-biotherapeutic system to continuously deliver L-DOPA stably, thereby relieving motor symptoms. Our data demonstrate that our engineered plasmid-based L-DOPA-expressing E. coli Nissle 1917 probiotic strain (EcN2LDOPA-P3) efficiently produced up to 12,000 ng/mL L-DOPA in vitro. In mouse model systems, EcN2LDOPA-P3 readily colonized for up to 48 h, achieved steady-state plasma L-DOPA concentrations, and increased brain L-DOPA and dopamine levels by 1- to 2-fold. Lastly, EcN2LDOPA-P3 significantly diminished motor and nonmotor behavioral deficits in a mouse model of PD compared to traditional chemical L-DOPA therapy. These findings support the therapeutic feasibility of a noninvasive, orally administered bioengineered bacterial therapy for the chronic delivery of L-DOPA, which may address limitations associated with current treatment alternatives.

|
Scooped by
mhryu@live.com
February 2, 11:36 PM
|
The search for new antimicrobial natural products from microorganisms has been limited by the transcriptional silencing of biosynthetic genes when microbes are cultivated outside their ecological environments. Nevertheless, applying knowledge of the ecological roles, for example, microbial defense against plant pathogens, can improve drug discovery efforts. Interactions between plants and their microbiota, during adaptation to pathogen stress, provide ecological cues that induce microenvironments suppressive to pathogens. This article highlights research linking pathogen-induced plant stress signals to the activation of microbial natural product biosynthesis, emphasizing the need for further studies on how plant metabolites can influence biosynthesis in plant-associated microbes.

|
Scooped by
mhryu@live.com
February 2, 10:43 PM
|
Fusarium oxysporum plays crucial roles as both a damaging plant pathogen and a model for studying host-pathogen interactions. It is well established that systematic functional genomics approaches are vital for advancing agricultural biotechnology and developing biological agents. Creating efficient gene-editing systems can help elucidate virulence mechanisms and enable rapid production of modified strains for practical use. However, current transformation methods face major challenges, such as high enzyme costs, and CRISPR/Cas9 systems are limited by PAM sequence availability and by blunt-end DNA cleavage, which hampers homologous recombination. Furthermore, complex guide RNA scaffolds complicate large-scale functional studies with traditional methods. To address these challenges, we have developed a streamlined CRISPR/Cpf1 (Cas12a) platform that combines small-scale protoplast preparation with significantly reduced enzyme use, exploiting Cpf1's unique features, such as flexible PAM recognition, staggered DNA cuts that promote recombination, improved target specificity, and simpler guide RNA design. This platform can also accelerate the development of biological agents and support high-throughput screening applications essential to the progress of agricultural biotechnology.

|
Scooped by
mhryu@live.com
February 2, 7:19 PM
|
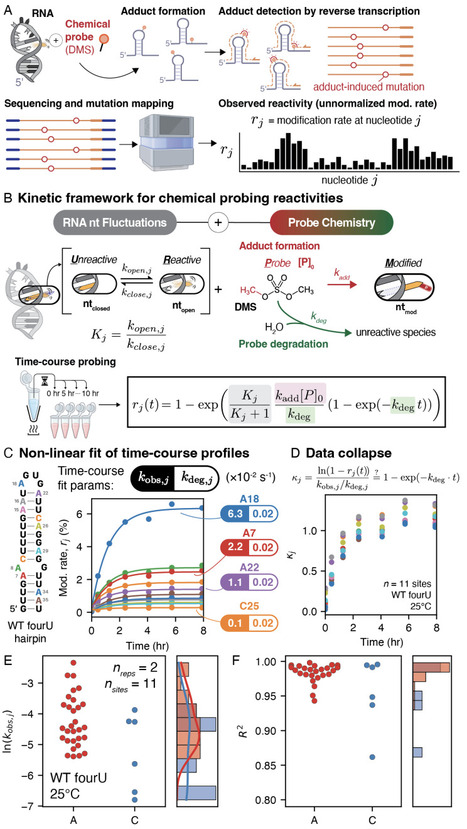
RNA function is governed by RNA folding, but strategies for measuring RNA folding thermodynamics are limited, and fundamental questions such as the energy of base pair opening remain debated. Here we introduce Probing-Resolved Inference of Molecular Energetics (PRIME) to extract nucleotide-resolution RNA structural energetics from scalable chemical probing experiments. Applying PRIME to diverse RNAs, we find that RNA base pairs and tertiary interactions dynamically open with free energies of 0.5-3 kcal/mol, revealing that RNA nucleotides ubiquitously sample open conformations at biologically accessible energies. PRIME further resolves energetic coupling across RNAs, providing an energetic understanding of RNA structural dynamics and long-range coordination in RNA folding. PRIME represents a widely accessible strategy for interrogating RNA thermodynamics, enabling mechanistic understanding and engineering of RNA biology.

|
Scooped by
mhryu@live.com
February 2, 4:54 PM
|
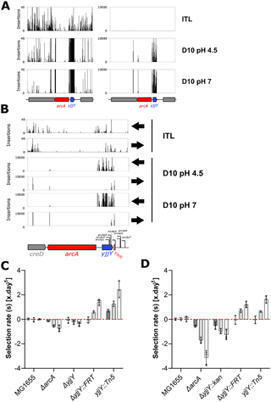
Long term laboratory evolution experiments are a powerful tool that are increasingly being used to study fundamental aspects of evolution and to identify genes that contribute to overall fitness under different conditions. However, even with automation, the time that they take to execute limits the extent to which evolution experiments can be used as part of a high throughput approach to understand the links between genotype and phenotype. Mutations that lead to genetic loss of function (LoF) are frequently selected for in evolution experiments. Thus, in principle these experiments could be done more rapidly by starting not with clonal isolates but with dense transposon libraries that will contain loss of function mutations in all non-essential genes. Here, we test this hypothesis by comparing the results of long term (5 month) evolution experiment, in which E. coli was grown with daily transfers in unbuffered LB starting at pH 4.5, with short term (5 and 10 day) experiments on a high-density transposon library in the same strain and under the same conditions. We show that there is a overlap in the genes and pathways identified using the two methods, as well as identifying other gene of interest whose LoF contributes to fitness. This approach has the potential to complement laboratory-based evolution, enabling rapid, higher throughput, testing of a wide range of parameters that may have an influence on evolutionary trajectories.

|
Scooped by
mhryu@live.com
February 2, 4:33 PM
|
Synthetic biology is transforming how we understand and teach microbial energy metabolism. In a recent study (F. Li, B. Zhang, X. Long, H. Yu, et al., Nat Commun 16:2882, 2025, https://doi.org/10.1038/s41467-025-57497-z), the authors demonstrated a synthetic gene circuit that enables Shewanella oneidensis to produce and release phenazine-1-carboxylic acid, a redox-active metabolite that enhances extracellular electron transfer and electricity generation. This perspective highlights the significance of their work, focusing on how controlling the production of redox mediators provides new insights into microbial electron flow and bioelectronic design. Beyond its technological implications, this system also serves as a valuable educational case study for teaching principles of redox balance, gene regulation, and metabolic engineering. Viewing this advancement in the context of biology education underscores the potential of synthetic circuits to deepen our understanding of microbial metabolism and to promote interdisciplinary learning in microbiology, biotechnology, and engineering.
|
 Your new post is loading...
Your new post is loading...