 Your new post is loading...

|
Scooped by
mhryu@live.com
January 16, 3:27 PM
|
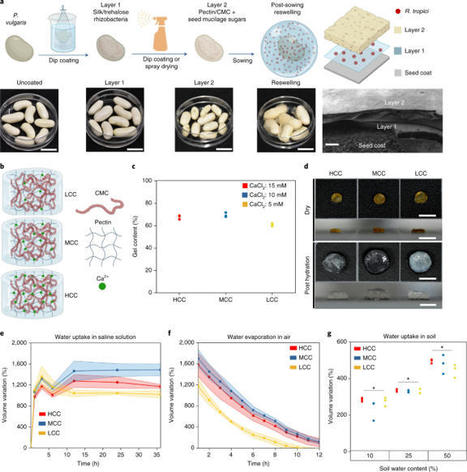
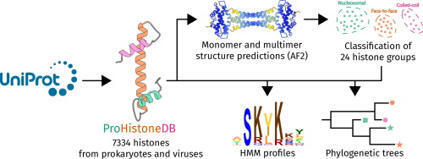
Histones are one of the fundamental chromatin proteins of life. In eukaryotes and megaviruses, they form nucleosome structures that wrap DNA. However, in prokaryotes, histones are much more diverse in how they organize DNA. In bacteria, histones bend and wrap DNA while in archaea they wrap and bridge DNA. These differences in DNA organizing properties are primarily due to distinct modes of histone multimerization. Here we present ProHistoneDB, an online database describing and categorizing prokaryotic and viral histones. For each histone, monomer, dimer, tetramer, and hexamer predictions are viewable and downloadable. ProHistoneDB contains 7334 histones, categorized into 24 groups based on the multimer predictions. For each category, interactive phylogenetic trees and HMM profile logos are available to identify conserved residues and explore the relative evolutionary relationships of histones. ProHistoneDB can be accessed at https://prohistonedb.org/.

|
Scooped by
mhryu@live.com
January 16, 2:47 PM
|
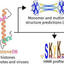
The legume-rhizobium symbiosis is a cornerstone of sustainable agriculture due to its ability to facilitate biological nitrogen fixation. Still, real-time visualization and quantification of this interaction remain technically challenging, especially across different host backgrounds. In this study, we systematically evaluate the efficacy of the nitrogenase system nifH promoter (PnifH) in driving expression of distinct fluorescent reporters; superfolder yellow fluorescent protein (sfYFP), superfolder cyan fluorescent protein (sfCFP), and various red fluorescent proteins (RFPs) within root nodules of determinate (Lotus japonicus-Mesorhizobium japonicum) and indeterminate (Pisum sativum-Rhizobium leguminosarum) systems. We show that PnifH-driven sfYFP and sfCFP yield strong, uniform, and reproducible fluorescence in nodules of both systems, facilitating reliable quantification of nodulation traits and strain occupancy. In contrast, RFPs including monomeric (mScarlet-I, mRFP1, mARs1) and multimeric (AzamiRed1.0) variants exhibited weak or inconsistent signals in pea. Notably, fluorescent labeling did not impair rhizobial competitiveness for root nodule occupancy, and PnifH-driven sfYFP and sfCFP reporters enabled robust multiplexed imaging in single-root and split-root assays. In the lotus, mScarlet-I worked robustly and facilitated a tripartite strain labeling system. Complementing our molecular toolkit, we established a deep learning-based analytical pipeline for high-throughput, automated quantification of nodulation traits, validated against standard ImageJ analysis. Altogether, our results identify PnifH-driven sfYFP and sfCFP as robust, broadly applicable reporters for legume-rhizobium symbiosis studies, while highlighting the need for optimized red fluorophores in some contexts. The integration of validated promoter-reporter constructs with state-of-the-art computational approaches provides a scalable framework for dissecting the spatial and competitive dynamics of plant-microbe mutualisms.

|
Scooped by
mhryu@live.com
January 16, 2:38 PM
|
Could codon composition condition the immediate success and the orientation of horizontal gene transfer? Horizontal gene transfer represents a change in the genome of expression of the transferred gene, and experimental evidence has accumulated indicating that the codon composition of a sequence is an important determinant of its compatibility with the translation machinery of the genome in which it is expressed. This suggests that codon composition influences the phenotype and the fitness conferred by a transferred gene and thus the immediate success of the transfer. To directly test this hypothesis, we characterized the resistance conferred by synonymous variants of a gentamicin resistance gene in three bacterial species: Escherichia coli, Acinetobacter baylyi and Pseudomonas aeruginosa. The strongest determinant of the resistance level conferred was the species in which the resistance gene was transferred, very likely because of important differences in the copy number of the plasmid carrying the gene. Significant differences in resistance were also found between synonymous variants within each of the three species, but more importantly, there was a strong interaction between species and variant: variants conferring high resistance in one species confer low resistance in another. However, the similarity in codon usage between the synonymous variants and the host genome only explained part of the phenotypic differences between variants in one species, P. aeruginosa. Further investigation of alternative explanations did not reveal common universal mechanisms across our three bacterial species. We conclude that codon composition can be a determinant of post-horizontal gene transfer success. However, there are multiple paths leading from synonymous sequence to phenotype, and sensitivity to these different paths is species-specific.

|
Scooped by
mhryu@live.com
January 16, 2:09 PM
|
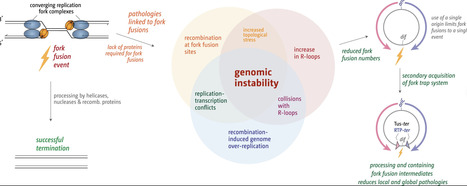
Termination of DNA replication is a surprisingly complex process that contributes critically to genome stability and cell viability. And even though progress was made to establish the consequences that arise if termination is going awry, the precise molecular mechanisms of fork fusion events and the coordination with key factors that ensure that DNA replication is brought to a successful conclusion remain poorly understood. We therefore investigated replication termination in E. coli, focusing specifically on the interplay between replication fork fusions and genomic stability, the Tus–ter replication fork trap, and key DNA-processing enzymes. By utilizing whole genome sequencing, immunoblotting, and recombination reporter assays, we demonstrate that local hyper-recombination is induced wherever forks meet and that the combined loss of factors such as RecG helicase and 3′ exonucleases causes extreme over-replication in the terminus region of the chromosome. Unexpectedly, cells lacking Tus exhibit elevated R-loop levels, revealing an unanticipated connection between the fork trap and R-loop metabolism. These findings underscore the complexity of replication termination and its central role in maintaining bacterial genome stability, while providing mechanistic insights with implications for understanding replication termination in more complex organisms and developing new antimicrobial strategies.

|
Scooped by
mhryu@live.com
January 16, 1:57 PM
|
We have evaluated the prediction accuracy of three different tools, deep-learning-based AlphaFold2, AlphaFold3, and large language model-based ESMFold, utilizing the experimentally derived structures deposited in the Protein Data Bank between 2022 and 2024, excluding those entries with close homologs in the structures released prior to 2022. Based on the criteria of sequence identity lower than 40% and query coverage <70%, 1666 monomeric and 994 dimeric proteins were selected as challenging targets for benchmarking. Our analysis showed that AlphaFold2 and AlphaFold3 correctly predicted 88% of monomeric structures and 77% of dimeric proteins. On the other hand, ESMFold accurately predicted 76% of the monomeric proteins and 41% of the dimeric proteins. Since most incorrect predictions involved nuclear magnetic resonance structures, benchmarking on X-ray and cryo-electron microscopy structures showed that the prediction accuracy of AlphaFold and ESMFold was 95% and 83%, respectively, for monomeric proteins. Overall, these findings demonstrate significant differences in the prediction accuracy of these machine learning (ML)-based tools for monomeric and dimeric proteins, highlighting the advantages and limitations of these tools. Finally, to facilitate easy access to benchmarking data, we developed ProModEv (Protein Model Evaluation portal), an interactive web portal for systematic analysis of these benchmarking results, and it is available at http://pdbi.nii.ac.in/ProModEv/.

|
Scooped by
mhryu@live.com
January 16, 1:24 PM
|
The canonical PAM site TTTV (where V = A, G, or C) is widely used in the design of CRISPR-Cas12a systems for both genome editing and diagnostic applications. Although several non-canonical protospacer-adjacent motifs (PAM) have been identified, they generally exhibit weak Cas12a cleavage activity. In this study, we find that increasing the reaction temperature to 45 °C or higher allows the identification of numerous non-canonical PAMs with trans-cleavage activity comparable to that of canonical PAMs, while displaying only weak cis-cleavage activity. Moreover, we observe that combining these non-canonical PAMs with elevated temperatures significantly enhances the Cas12a system’s ability to discriminate highly similar sequences. Based on these findings, we develop a non-canonical PAM-mediated, poikilothermal, one-pot CRISPR-Cas12a detection platform (POP-CRISPR), which demonstrates substantial improvements in sensitivity, specificity, speed, and target adaptability for nucleic acid detection compared to existing methods. These advantages are validated through the reliable detection of clinical samples, including those of Human papillomavirus (HPV), Mycoplasma pneumoniae (MP), and its drug-resistant strains. Additionally, we show that POP-CRISPR enables rapid, on-site pathogen detection within 20 min, using a fast sample processing protocol and a miniaturized detection device. CRISPR-Cas12a diagnostics are limited by strict PAM requirements. Here, authors show that higher temperatures activate numerous noncanonical PAMs, enabling a versatile one-pot platform that improves sensitivity, specificity, and rapid on-site pathogen detection.

|
Scooped by
mhryu@live.com
January 16, 12:38 PM
|
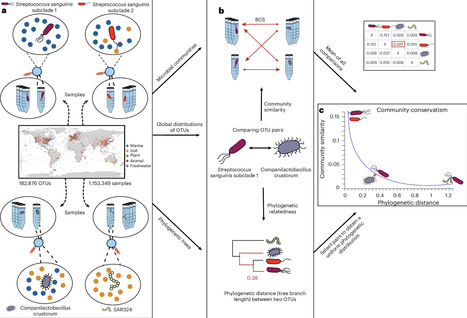
Phylogenetic signal describes the tendency of related organisms to resemble each other in morphology and function. Related organisms tend to also live in similar ecological niches, which is termed niche conservatism. The concepts of both phylogenetic signal and niche conservatism are widely used to understand crucial aspects of evolution and speciation, and they are well established in animals and plants. However, although assumed to be present, the extension of these concepts to microorganisms is challenging to assess. Here we hypothesize that two closely related microbial species should be found in samples with similar community compositions, reflecting their ecological similarity. We propose ‘community conservatism’ to refer to this phenomenon and leverage a database with millions of samples and hundreds of thousands of pairs of microorganisms to assess their relatedness and the similarity of the communities they occupy. Our findings reveal that community conservatism can be observed globally in all environments and phyla tested, over nearly all taxonomic ranks, but to varying extents. Analyzing community conservatism shows promise to advance our understanding of evolution, speciation and the mechanisms governing community assembly in microorganisms. Furthermore, we propose that it can be used to reintegrate ecological parameters into operational taxonomic unit delimitation. This study reveals that closely related microorganisms tend to inhabit similar communities across all major environments and phyla. The authors term this phenomenon ‘community conservatism’, extending the ecological concepts of phylogenetic signal and niche conservatism to the microbial world.

|
Scooped by
mhryu@live.com
January 16, 9:45 AM
|
Antibiotic resistance is arguably one of the greatest threats to global health today. The worldwide emergence of multidrug-resistant and hypervirulent Klebsiella pneumoniae underscores the urgent need for alternative treatments. Bacteriophages (phages) are considered one of the most promising alternatives to address this crisis. In this review, we summarize current knowledge of phage–host interactions and highlight recent advances in phage therapy against K. pneumoniae, including phage cocktails, antibiotic combination therapy, and treatments based on phage-derived proteins. Despite their tremendous therapeutic potential, significant challenges remain. We therefore also discuss strategies to optimize phage research and recent innovations in the field.

|
Scooped by
mhryu@live.com
January 16, 12:41 AM
|
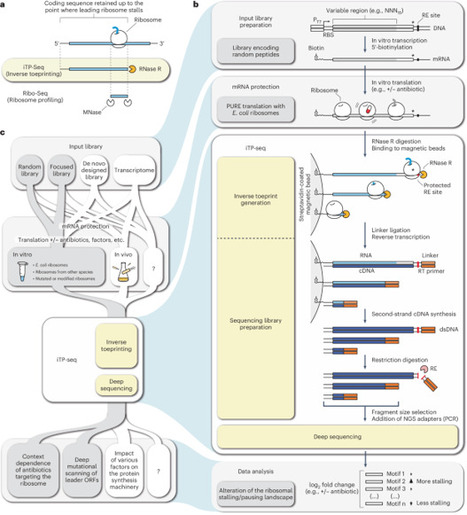
Uneven translation rates resulting from mRNA context, tRNA abundance, nascent amino acid sequence or various external factors play a key role in controlling the expression level and folding of the proteome. Inverse toeprinting coupled to next-generation sequencing (iTP-seq) is a scalable in vitro method for characterizing bacterial translation landscapes, complementary to ribosome profiling (Ribo-seq), a widely used method for determining transcriptome-wide protein synthesis rates in vivo. In iTP-seq, ribosome-protected mRNA fragments known as inverse toeprints are generated by using RNase R, a highly processive 3′ to 5′ RNA exonuclease. Deep sequencing of these fragments reveals the position of the leading ribosome on each mRNA with codon resolution, as well as the full upstream coding regions translated by these ribosomes. Consequently, the method requires no a priori knowledge of the translated sequences, enabling work with fully customizable transcript libraries rather than previously sequenced genomes. As a standardized framework for inverse toeprint generation, amplification and sequencing, iTP-seq can be used in combination with different types of libraries, in vitro translation conditions and data-analysis pipelines tailored to address a range of biological questions. Here, we present a robust protocol for iTP-seq and show how it can be integrated into a broader workflow to enable the study of context-dependent translation inhibitors, such as antibiotics. The time required to complete this workflow is ~10 d, and the workflow can be carried out by an experienced molecular biologist, with data analysis also requiring a working knowledge of command-line tools and Python scripts. This protocol describes inverse toeprinting coupled to next-generation sequencing, an in vitro approach to characterize bacterial translation at codon resolution that can accommodate custom synthetic libraries and various translation perturbations.

|
Scooped by
mhryu@live.com
January 15, 11:47 PM
|
Recirculating aquaculture system (RAS) effluents contain substantial nitrate and phosphate loads, posing a significant eutrophication risk to aquatic ecosystems if left untreated. While microalgal bioremediation is a promising strategy, expanding opportunities for valorisation is crucial for its successful implementation. This study couples the cultivation of nitrate-utilizing Chlamydomonas reinhardtii strain CC-1690 in RAS effluent with photobiological hydrogen (H2) production to achieve nutrient removal, bioenergy production, and high-quality biomass. Within 70 h, CC-1690 achieved near-complete depletion of nitrate and phosphate from the effluent. Subsequently, H2 production was induced via carbon limitation and anoxia, yielding a cumulative 49 umol H2 mg−1 Chl over 72 h. Although decreased chlorophyll content and Fv/Fm indicated physiological stress, key photosynthetic subunits (PsbA, PSBO, CP47, PetB and PsaA) remained largely stable. Importantly, the process preserved biomass quality; total lipid content increased slightly, enriched in palmitic (16:0) and α-linolenic (18:3ω-3) fatty acids while protein content was unaffected. These results demonstrate that integrating H2 production with RAS effluent bioremediation offers a robust circular economy approach, valorising wasted nutrients into bioenergy and high-quality biomass with potential downstream applications.

|
Scooped by
mhryu@live.com
January 15, 11:25 PM
|
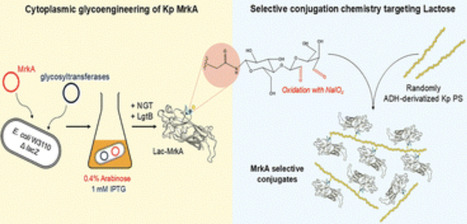
Chemical conjugation to carrier proteins has been traditionally used to improve polysaccharides immunogenicity and to overcome the limitations of T-independent antigens, including lack of immunological memory and efficacy in infants. A double-hit approach, meaning that both polysaccharide and carrier protein belong to the same pathogen, may be particularly useful for targeting bacterial species with large glycan variability. Recently, bacterial protein glycosylation has been exploited to obtain glycosylated proteins in E. coli cytoplasm. In our work we have combined cytoplasmic glycoengineering and chemical conjugation for the development of novel selective glycoconjugates, with the aim to preserve the immunogenicity of the protein chosen as carrier. The potential protective protein MrkA, the major component of Klebsiella pneumoniae type 3 fimbriae, was successfully modified with a lactose moiety in E. coli. K. pneumoniae K2 K-antigen and O1v1 O-antigen were then covalently linked to MrkA at the level of this unique sugar handle and tested in vivo. Immune response against MrkA and sugars was evaluated in animal models. This work contributes to expand the application of the glycoengineering technology for the development of effective glycoconjugate vaccines.

|
Scooped by
mhryu@live.com
January 15, 11:16 PM
|
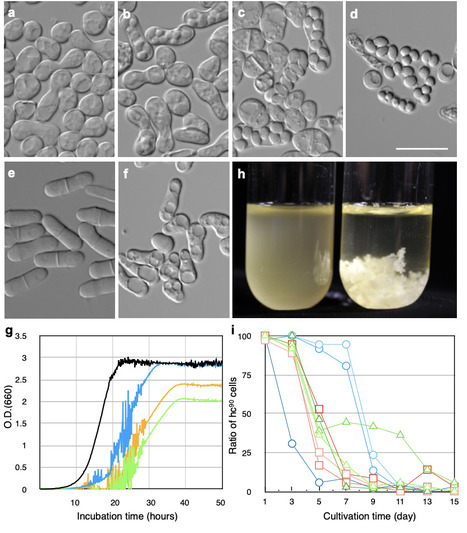
Sexual reproduction in yeast typically occurs under nutrient-limited conditions, producing stress-resistant spores that promote survival. However, wild Schizosaccharomyces japonicus strains isolated from fruit flies exhibit constitutive sporulation even under nutrient-rich conditions. Here we demonstrate that this nutrient-independent sporulation enhances survival by enabling spores to resist digestion by Drosophila melanogaster, and that although such strains are disadvantageous under nutrient-rich conditions, they are positively selected during insect predation. Genetic analyses reveal that multiple mutations in six meiotic regulatory genes, acting via epistasis, underlie this phenotype. This trait is widely distributed across natural populations throughout Japan. These findings suggest that constitutive sexual reproduction functions as an adaptive strategy facilitating insect-mediated dispersal, reflecting an ecological context where sexual reproduction is maintained despite its costs. This work provides new insights into yeast-insect symbiosis, evolutionary dynamics of sexual adaptation, and potential applications in yeast breeding.

|
Scooped by
mhryu@live.com
January 15, 8:31 PM
|
Understanding how cells make decisions over time requires the ability to link past molecular states to future phenotypic outcomes. We present TimeVault, a genetically encoded system that records and stores transcriptomes within living mammalian cells for future readout. TimeVault leverages engineered vault particles that capture mRNA through poly(A) binding protein. We demonstrate that the transcriptome stored by TimeVaults is stable in living cells for over 7 days. TimeVault enables high-fidelity transcriptome-wide recording with minimal cellular perturbation, capturing transient stress responses and revealing gene expression changes underlying drug-naive persister states in lung cancer cells that evade EGFR inhibition. By linking past and present cellular states, TimeVault provides a powerful tool for decoding how cells respond to stress, make fate decisions, and resist therapy.
|

|
Scooped by
mhryu@live.com
January 16, 2:52 PM
|
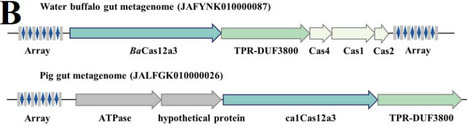
The CRISPR-Cas12 family encompasses diverse RNA-guided nucleases with both DNA-targeting and RNA-targeting subtypes. They can trigger antiviral activities mainly through either direct elimination invading nucleic acids, or activating broad collateral cleavage to induce abortive infection. Here, we report a novel type V CRISPR effector BaCas12a3 that causes growth inhibition through a unique tRNA-cleavage mechanism. Plasmid interference assays indicated that BaCas12a3 inhibits host growth arrest without invoking the DNA damage response, suggesting that the immune responses may not involve double strand breaks of DNA. Indeed, biochemical characterization of the BaCas12a3-crRNA ribonucleoprotein (RNP) unraveled that the effector is an RNA-activating nuclease that cleaves 3′ terminus of tRNAs. Cryo-EM structures of BaCas12a3 reveal a conserved bilobed architecture featuring a unique nucleic acid-loading (NL) domain adjacent to the RuvC catalytic center. Structural and mutagenesis analyses show that the NL domain, together with a zinc ribbon domain, form a gated substrate groove. Target RNA binding induces conformational changes that open this groove and expose the RuvC active site, enabling specific tRNA cleavage while preventing other non-specific degradation. Our findings identified the NL domain aside the RuvC active site responsible for the tRNA recognition in BaCas12a3, expanding the functional diversity of CRISPR immunity.

|
Scooped by
mhryu@live.com
January 16, 2:43 PM
|
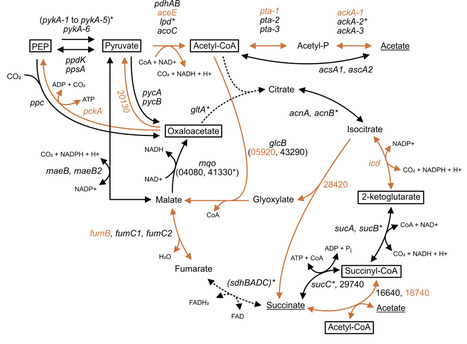
Nitrogen-fixing microbes are a primary contributor of this important nutrient to the global nitrogen cycle. Biological nitrogen fixation (BNF) through the enzyme nitrogenase requires extensive energy that in whole cells is generally studied during the oxidation of carbohydrates such as sugars. The nitrogen-fixing bacterium Azotobacter vinelandii is a model diazotroph for the study of aerobic BNF. Much is known about metabolism in A. vinelandii when cultured on a simple medium where energy is provided primarily in the form of sucrose or glucose. Outside of the laboratory, this soil bacterium grows on metabolites primarily derived from plant root exudates or from the degradation of dead plant matter. In this work, we expand on previous studies looking at genes that are essential to BNF in A. vinelandii when grown on sucrose medium using transposon sequencing (Tn-seq). We applied Tn-seq to determine the genes essential to growth when the medium was shifted to acetate, succinate or glycerol as the primary carbon and energy source to fuel both growth and BNF. A global overview of the genes of central metabolism and those directing substrates toward central metabolism, along with a selection of unexpected genes that were essential for specific growth substrates, is provided.

|
Scooped by
mhryu@live.com
January 16, 2:34 PM
|
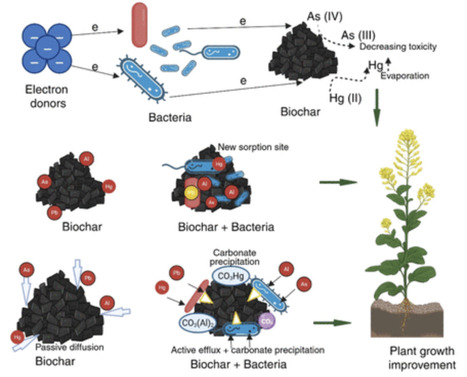
Soil microorganisms are essential drivers of ecosystem functioning and mediate pollutant degradation, metal detoxification, and nutrient cycling. This review aims to synthesize recent mechanistic advances in understanding how microbes degrade organic contaminants, transform or immobilize metals, mitigate toxic effects on plants through chelation, redox reactions, sequestration, and support soil structure and fertility. Microbial consortia and rhizosphere-associated taxa accelerate pollutant breakdown, reduce metal toxicity, and enhance plant resilience in acidic or contaminated soils. Integration of microbial processes with amendments such as biochar and organic matter further improve remediation efficiency and sustainability. Key insights reveal that microbial signaling networks, biofilm formation, and plant–microbe interactions are critical for maintaining the ecosystem stability under stress. These findings underscore the potential of microbial driven strategies to restore degraded soils, minimize reliance on chemical inputs, and promote sustainable agricultural practices, although field-scale persistence and ecological interactions warrant further research.

|
Scooped by
mhryu@live.com
January 16, 2:00 PM
|
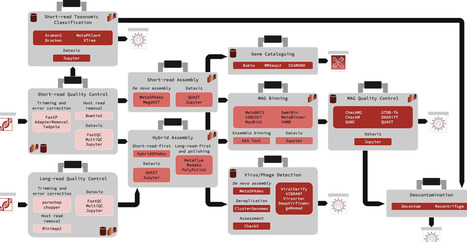
Computational analysis of large-scale metagenomics sequencing datasets provides valuable isolate-level taxonomic and functional insights from complex microbial communities. However, the ever-expanding ecosystem of metagenomics-specific methods and file formats makes designing scalable workflows and seamlessly exploring output data increasingly challenging. Although one-click bioinformatics pipelines can help organize these tools into workflows, they face compatibility and maintainability challenges that can prevent replication. To address the gap in easily extensible yet robustly distributable metagenomics workflows, we have developed the Core Analysis Modular Pipeline (CAMP), a module-based metagenomics analysis system written in Snakemake, with a standardized module and directory architecture. Each module can run independently or in sequence to produce target data formats (e.g. short-read preprocessing alone or followed by de novo assembly), and provides output summary statistics reports and Jupyter notebook-based visualizations. We applied CAMP to a set of 10 metagenomics samples, demonstrating how a modular analysis system with built-in data visualization facilitates rich seamless communication between outputs from different analytical purposes. The CAMP ecosystem (module template and analysis modules) can be found at https://github.com/Meta-CAMP.

|
Scooped by
mhryu@live.com
January 16, 1:35 PM
|
Yeast biosensors represent a promising biotechnological innovation for ensuring the safety and quality of fermented beverages such as beer, wine, and kombucha. These biosensors employ genetically engineered yeast strains to detect specific contaminants, spoilage organisms, or hazardous compounds during fermentation or the final product. By integrating synthetic biology tools, researchers have developed yeast strains that can sense and respond to the presence of heavy metals (e.g., lead or arsenic), mycotoxins, ethanol levels, or unwanted microbial metabolites. When a target compound is detected, the biosensor yeast activates a reporter system, such as fluorescence, color change, or electrical signal, providing a rapid, visible, and cost-effective means of monitoring safety parameters. These biosensors offer several advantages: they can operate in real time, are relatively low-cost compared to conventional chemical analysis methods, and can be integrated directly into the fermentation system. Furthermore, as Saccharomyces cerevisiae is generally recognized as safe (GRAS), its use as a sensing platform aligns well with existing practices in beverage production. Yeast biosensors are being investigated for the early detection of contamination by spoilage microbes, such as Brettanomyces and lactic acid bacteria. These contaminants can alter the flavor profile and shorten the product’s shelf life. By providing timely feedback, these biosensor systems allow producers to intervene early, thereby reducing waste and enhancing consumer safety. In this work, we review the development and application of yeast-based biosensors as potential safeguards in fermented beverage production, with the overarching goal of contributing to the manufacture of safer and higher-quality products. Nevertheless, despite their substantial conceptual promise and encouraging experimental results, yeast biosensors remain confined mainly to laboratory-scale studies. A clear gap persists between their demonstrated potential and widespread industrial implementation, underscoring the need for further research focused on robustness, scalability, and regulatory integration.

|
Scooped by
mhryu@live.com
January 16, 1:05 PM
|
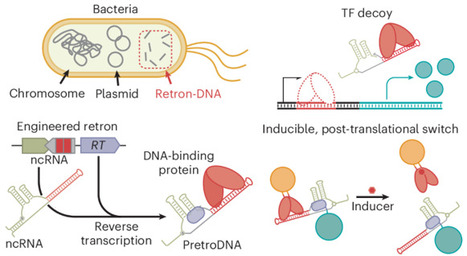
Life’s development and adaption to fluctuating environments are underpinned by DNA–protein interactions, which have also spurred the development of artificial systems ranging from genetic circuits to nanoarchitectures. Nonetheless, in cellulo there remains an untapped design space for DNA–protein systems that are incompatible with the role of DNA as genetic material. Here we engineer retrons to intracellularly express non-genetic small DNAs embedding specific protein-binding sequences. These DNA species are products of genes and therefore their quantitative, spatial and functional control is decoupled from genetic stability, enabling alternative architectures of DNA–protein systems with unique functions. Using synthetic networks of proteins and the engineered retron-DNA, we demonstrated precise, multiplexed gene regulation and construction of feedback circuits for dynamic responses. Further, we developed DNA-based molecular scaffolds and bridges that enable modular, post-translational and spatial control of multiple proteins within cells. Finally, we transformed an allosteric transcription factor into inducible post-translational switches. Our work suggests that the non-genetic DNA–protein systems represent a promising control layer for creating synthetic cellular behaviours. The design space of synthetic DNA–protein systems in cells has been constrained by DNA’s role as genetic material. Now it has been shown that engineering retron-derived ‘non-genetic’ small DNAs for sequence-specific protein binding enables alternative DNA–protein networks for gene regulation, feedback loops, molecular scaffolds and post-translational switches.

|
Scooped by
mhryu@live.com
January 16, 10:38 AM
|
This research utilized the CRISPR/Cas9 editing method to generate six mutant strains of Escherichia coli (E. coli) DH5α targeting specific genes. The functional characterization and phenotypic analysis confirmed the regulatory roles of these genes in modifying membrane permeability. The variations in membrane permeability among the mutant strains were assessed by measuring electrical conductivity, ortho-nitrophenyl-β-D-galactopyranoside (ONPG) hydrolysis, and propidium iodide (PI) fluorescence, with E. coli DH5α:ompA′ exhibiting the most pronounced increase in membrane permeability. The function of these genes in transformation was analyzed from physicochemical and microscopic perspectives. Assays of plasmid transformation efficiency revealed a significant enhancement in the E. coli DH5α:ompA′ mutant strain, underscoring the critical function of outer membrane proteins in DNA acquisition. Permeability simulations were performed utilizing the E. coli DH5α:ompA′ mutant strain, grounded in a previously established model. The quantitative correlation between transformation efficiency and membrane permeability in this mutant conformed to the equation T = aP + c.

|
Scooped by
mhryu@live.com
January 16, 12:45 AM
|
Phage genome engineering methods accelerate the study of phage biology, the discovery of new functions, and the development of innovative genetic engineering tools. Here, we present QuickPhage, a rapid, technically accessible, precise, and cost-effective method for engineering Bacillus subtilis phages. Our approach uses CRISPR-Cas9 as a counter-selection system to isolate mutants of the model lytic siphovirus phage, SPP1. Efficient genome editing was achieved using homologous repair patches as short as 40 nucleotides, enabling streamlined patch construction and parallel engineering, resulting in highly accurate genome edits within a day. We applied QuickPhage to delete both essential and nonessential phage genes and to insert reporter genes. Protein production, such as GFP, was synthetically regulated using inducible systems without significantly affecting phage fitness, achieving induction levels of up to 400-fold. Time-series coinfection experiments with fluorescent protein expressing phages also revealed a highly efficient superinfection arrest mechanism that prevents reinfection as early as 13 min after initial infection. These findings highlight the potential of phages for protein production, opening new opportunities for metabolic engineering. This work also lays the foundation for systematic phage genome refactoring workflows and the development of phage-based tools for efficient DNA delivery, thereby expanding the synthetic biology toolbox for B. subtilis.

|
Scooped by
mhryu@live.com
January 16, 12:09 AM
|
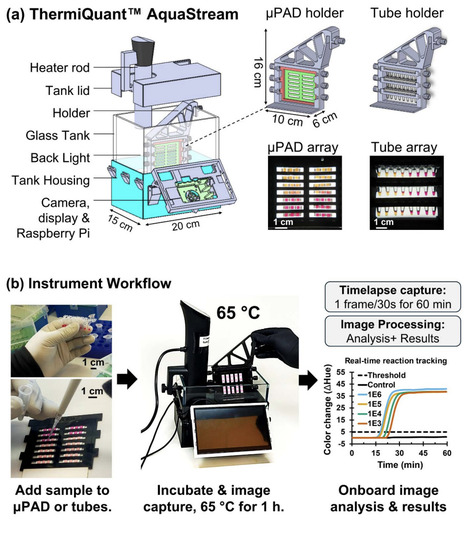
Microfluidic paper-based analytical devices (uPADs) are an attractive format for colorimetric nucleic acid amplification tests (NAATs) because they enable low-cost, portable diagnostics in resource-limited settings. However, researchers often optimize assays in liquid reactions in tubes before translating them to uPADs. Since both formats require separate instruments for incubation and real-time sensing, direct comparison of reactions between the two formats is difficult. To address these cross-platform limitations, we developed ThermiQuantTM AquaStream, a portable benchtop device (15 x 20 x 16 cm, ~5 kg; cost: USD 327) that supports seamless colorimetric loop-mediated isothermal amplification (LAMP) reactions in both uPADs and tubes under a common workflow. The system enables real-time reaction tracking (every 30 seconds) through onboard image processing, precise isothermal control (+/- 0.5 degree C) using a repurposed consumer-grade sous-vide heater, and medium-throughput (24 tubes or 42 uPADs). Testing with synthetic SARS-CoV-2 orf7ab DNA fragments demonstrated a limit of detection (LoD) of 50 copies per reaction in both formats (6.7 copies/uL for uPADs; 10 copies/uL for tubes). Standard calibration curves based on quantification time (Tq) and log10 DNA concentration showed strong full range linearity in tubes (from 1E6 to 50 copies/reaction, R2 = 0.98) and piecewise linearity in uPADs, with quantification reliability only above 1,000 copies/reaction (133 copies/uL, R2 = 0.99), which we set as the limit of quantification, LoQ. By unifying assay optimization across tube and uPAD formats, ThermiQuantTM AquaStream provides an affordable and versatile benchtop tool for molecular diagnostics for One Health applications in the clinic, on farms, and in the field.

|
Scooped by
mhryu@live.com
January 15, 11:36 PM
|
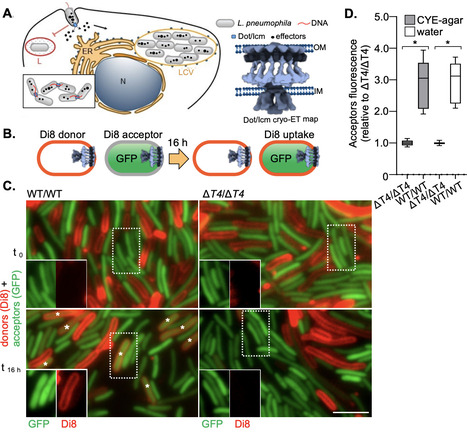
Type IV secretion systems (T4SS) are versatile molecular machines used by bacteria to secrete protein effectors into host cells, promoting pathogenesis, and to transfer DNA between bacteria through conjugation, driving horizontal gene transfer. Most, like Dot/Icm of the pathogen Legionella pneumophila or E. coli RK2, are primed for substrate delivery only upon contact with a target membrane, but mechanisms are unknown. A pilus could bind a receptor to initiate priming, but many T4SSs, especially those that deliver effectors, lack a pilus. Here, we present evidence that T4SSs are primed by direct contact with target membrane lipids. Combining fluorescence assays with genetics and biochemistry, we found that Dot/Icm drives lipid exchange between bacterial cells and between bacteria and synthetic membranes containing only lipids. Lipid exchange requires membrane contact but does not require ATP hydrolysis or even full complex assembly. Minimally, the outer membrane core complex protein DotG needs to be present in at least one of the apposed membranes. We similarly observed lipid mixing with the simpler E. coli RK2 T4SS, where we could follow lipid mixing and plasmid transfer simultaneously. We found that lipid mixing always preceded or accompanied plasmid transfer, suggesting it may be part of the contact-dependent priming mechanism. Lipid mixing was inhibited or promoted by lipids that inhibit or promote membrane fusion, respectively. Lipids inhibiting lipid mixing also inhibited substrate transfer. Together, our results suggest that initial contact between DotG outer segments and target membrane lipids promotes lipid mixing as part of the mechanism that primes T4SS for substrate translocation.

|
Scooped by
mhryu@live.com
January 15, 11:19 PM
|
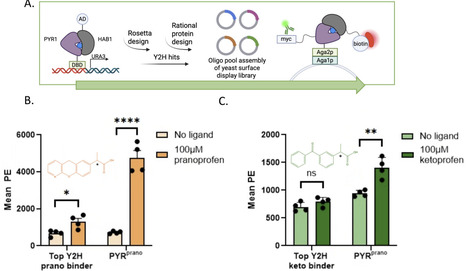
Non-steroidal anti-inflammatory drugs (NSAIDs) are pervasive environmental contaminants due to their frequent and widespread use, multiple paths of release into surface and ground water supply, diversity of the chemical class, and toxicity to aquatic and other non-target species. In particular, the 2-arylpropionic acid ("profen") class of NSAIDs poses significant risks to aquatic ecosystems due to incomplete removal during wastewater treatment. Current monitoring precludes high frequency testing at point sources. Here we present the engineering and application of a genetically encodable, protein-based biosensor for the detection of the NSAIDs ketoprofen and pranoprofen in wastewater effluent. We repurposed the plant hormone receptor PYR1 to bind selectively to profens using computational protein design, deep mutational scanning, and yeast 2 hybrid and yeast surface display screening. The resulting sensor, PYRNSAID, has a nanomolar limit of detection for ketoprofen and panoprofen, and micromolar sensitivity to the NSAIDs ibuprofen, fenoprofen, tolmetin and diclofenac. We also demonstrated dose responsive-activity of our sensor in simulated wastewater matrices containing the common wastewater contaminants sulfamethoxazole, caffeine, acetaminophen, and 2,4, dichlorophenol using a split Nanoluc luminescence assay. PYRNSAID is the first step towards a scalable, cost-effective alternative for real-time monitoring of pharmaceutical pollution.

|
Scooped by
mhryu@live.com
January 15, 8:46 PM
|
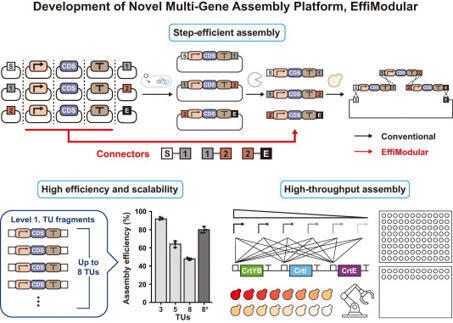
Rewiring the metabolic flux for efficient microbial conversion requires robust, scalable gene assembly. However, conventional gene assembly approaches are labor-intensive, highly experience-dependent, and require extensive expertise to ensure reproducibility and efficiency. Even with advanced automation platforms such as biofoundries, assembling gene arrays with multiple transcriptional units (TUs) remains challenging. In this study, we present Efficient Modular Gene Assembly (EffiModular), an integrated in vitro and in vivo gene assembly platform compatible with automated workflows. EffiModular enables the assembly of up to eight TUs with 80% efficiency in a single transformation. Integrated into a biofoundry workflow, it enabled the construction of 120 distinct yeast strains with varying levels of expression of the β-carotene biosynthesis genes within 3 days. Compared with conventional approaches, it significantly reduces procedural complexity, minimizes reliance on operator expertise, and accelerates workflow timelines. These features establish EffiModular as a next-generation gene assembly platform for scalable, reproducible gene assembly in biofoundry-based genetic engineering.
|
 Your new post is loading...
Your new post is loading...













marelli b.