 Your new post is loading...

|
Scooped by
?
Today, 2:49 PM
|
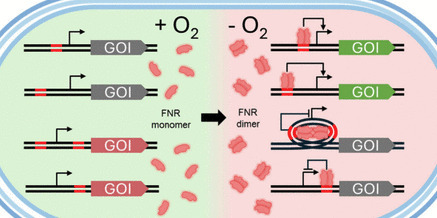
Promoters responsive to changes in cultivation conditions are essential tools for dynamic metabolic engineering. Oxygen-responsive promoters, in particular, exhibit significant application potential in oxygen-limited fermentation processes. However, currently reported oxygen-dependent promoters exhibit limited dynamic ranges, and notably, there remains a lack of research on oxygen-responsive negatively regulated promoters. In this study, we designed and characterized a series of dissolved oxygen-responsive promoters in Escherichia coli under the regulation of the transcription factor fumarate-nitrate reduction (FNR). Anaerobically activated promoters were constructed by inserting FNR binding site (FBS) upstream of inducible core promoters, while anaerobically repressed promoters were developed by inserting FBS downstream of or flanking constitutive promoters. The most effective anaerobically activated promoters showed 24–138-fold higher activity under anaerobic conditions compared to aerobic conditions. Under anaerobic conditions, promoters with DNA looping-mediated anaerobic repression maintained only 8–17% of the activity observed under aerobic conditions. These promoters were specifically regulated by FNR, as confirmed by tests in a DH5α Δfnr strain, and responded rapidly to oxygen depletion (within 30 min). The utility of these genetic tools was demonstrated by applying them to enhance pyruvate production in E. coli. An engineered strain with anaerobic-repressed aceE and anaerobic-activated atpAGD genes produced 5.76 g/L pyruvate at 55.7% yield in shake flask fermentations. This study offers an expanded toolbox of oxygen-responsive promoters that enable precise gene regulation based on dissolved oxygen levels, providing novel genetic strategies for developing efficient two-stage fermentation processes with separated growth and production phases.

|
Scooped by
?
Today, 2:44 PM
|
RNA design has emerged to play a crucial role in synthetic biology and therapeutics. Although tertiary structure-based RNA design methods have been developed recently, they still overlook the broader molecular context, such as interactions with proteins, ligands, DNA, or ions, limiting the accuracy and functionality of designed sequences. To address this challenge, we present RISoTTo (RIbonucleic acid Sequence design from TerTiary structure), a parameter-free geometric deep learning approach that generates RNA sequences conditioned on both their backbone scaffolds and the surrounding molecular context. We evaluate the designed sequences based on their native sequence recovery rate and further validate them by predicting their secondary structures in silico and comparing them to the corresponding native structures. RISoTTo performs well on both metrics, demonstrating its ability to generate accurate and structurally consistent RNA sequences. Additionally, we present an in silico design study of domain 1 of the NAD+ riboswitch, where RISoTTo generated sequences are predicted to exhibit enhanced binding affinity for both the U1A protein and the NAD+ ligand.

|
Scooped by
?
Today, 2:32 PM
|
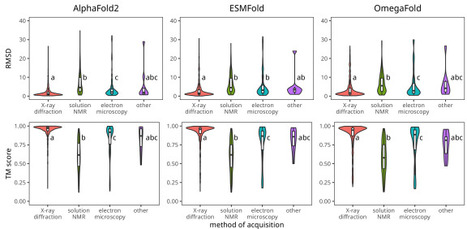
We compared the performance of three widely used protein structure prediction tools - AlphaFold2, ESMFold, and OmegaFold - using a dataset of over 1,300 newly created records from the PDB database. These structures, resolved between July 2022 and July 2024, ensure unbiased evaluation, as they were unavailable during the training of these tools. Using metrics such as root mean square deviation (RMSD), template modeling score (TM-score), and predicted local distance difference test (pLDDT), we found that AlphaFold2 consistently achieves the highest accuracy but depends on high-quality sequence alignments. In contrast, ESMFold and OmegaFold provide faster predictions and excel in challenging cases, such as rapidly evolving or designed proteins with limited sequence homology. Comparing ESMFold and OmegaFold, ESMFold achieves higher confidence scores (pLDDT) and structural similarity (TM-score). OmegaFold is competitive in specific contexts, such as de novo-designed proteins or sequences with limited evolutionary information. Additionally, we demonstrate that machine learning models trained on protein language model embeddings and pLDDT confidence scores can predict potential structure prediction failures, helping to identify challenging cases early in the pipeline.

|
Scooped by
?
Today, 2:25 PM
|
The scientific study of animal venoms covers a broad phylogenetic domain. We argue that the true extent of this domain has been obscured by researchers having overlooked the biological essence of venom. Venoms manipulate the physiological functioning of recipients to produce extended phenotypes that are beneficial to the venom producer and detrimental to its victim. The ability to produce extended phenotypes in living victims, such as prey paralysis, distinguishes venom from saliva. Understanding venom from this perspective substantially broadens the phylogenetic domain of venom to include taxa that use toxic secretions to feed on plants and manipulate sexual partners, and it paves the way for unifying the field of venomics with the fields that study invertebrate–plant interactions and sexual conflict

|
Scooped by
?
Today, 1:36 PM
|
Synthetic biology can endow microorganisms with novel capabilities. Su et al. developed an engineered strain of Vibrio natriegens that efficiently degrades complex organic pollutants in high-salinity environments. This technology offers a new biotechnological solution to urgent global environmental challenges, such as petrochemical wastewater discharges and marine oil spills.

|
Scooped by
?
Today, 1:28 PM
|
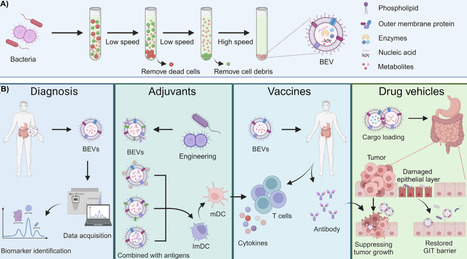
Bacterial extracellular vesicles (BEVs) are nanoscale spherical particles with lipid bilayer membranes containing diverse functional components from their parent bacteria. They exert pivotal effects on bacteria–bacteria and host–bacteria communication. Analyzing the dynamic changes in the production and composition of BEVs provides insights into the relationship between gut microbiota and host health, offering valuable perspectives for diagnosing various gastrointestinal diseases. Furthermore, BEVs can be employed as natural medications and drug delivery vehicles for inflammation management, cancer treatment, and vaccine development. This review summarizes the structural composition, generation mechanism, and biomedical applications of BEVs, emphasizing recent advances in immune regulation, gut microbiota modulation, and the clinical translation challenges associated with gastrointestinal diseases.

|
Scooped by
?
Today, 1:25 PM
|
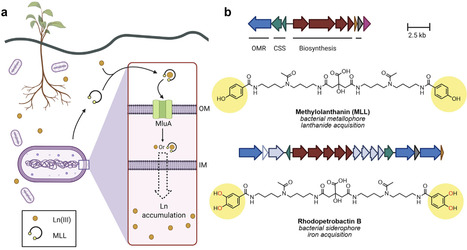
Metallophores are metal-chelating secondary metabolites that bacteria, fungi, and other organisms produce to acquire essential metal nutrients from their environment. This review highlights some recent discoveries in the field: the role of yersiniabactin in quorum sensing, the capacity of aspergillomarasmine A to inhibit bacterial Ni uptake, and the identification of methylolanthanin, a novel metallophore that enables bacterial lanthanide acquisition.

|
Scooped by
?
Today, 1:07 PM
|
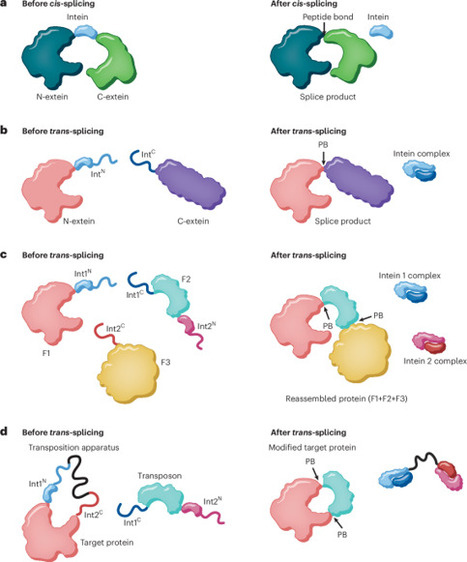
Transposition is normally considered a DNA-based process. Now, scientists have devised a means to carry out a similar transformation — this time using proteins. The approach enables protein semisynthesis and site-specific modification of folded substrates in vitro and in live cells, opening up new ways to probe and manipulate biological systems.

|
Scooped by
?
Today, 1:27 AM
|
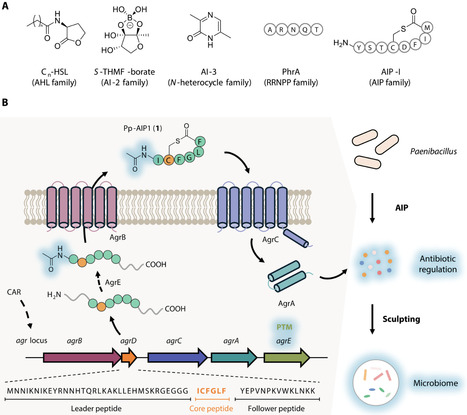
Microbes use signaling molecules to regulate multiple physiological processes and mediate chemical interactions. Decoding these chemical languages is instrumental in comprehending microbial regulatory mechanisms within complex microbiota. Here, we discover previously unidentified autoinducing peptides (AIPs) derived from the plant probiotic bacterium Paenibacillus polymyxa, identified as Pp-AIPs. Omics analyses coupled with genetic manipulations revealed that Pp-AIP1 could effectively modulate the production of multiple antimicrobial secondary metabolites at nanomolar concentration, expanding known AIP functions. Furthermore, through inoculating P. polymyxa in the natural rhizosphere microbiome and analyzing its antagonistic interactions against root microbes, we suggest that Pp-AIPs may influence the microbial community composition through modulating the antimicrobial spectrum. Global analysis of biosynthetic gene clusters (BGCs) reveal widespread co-occurrence of uncharacterized AIPs with secondary metabolite BGCs. This study underscores the unreported roles of AIPs in antibiotic regulation and the microbiome interactions, advancing knowledge of quorum-sensing mechanisms in microbial ecosystems.

|
Scooped by
?
Today, 12:59 AM
|
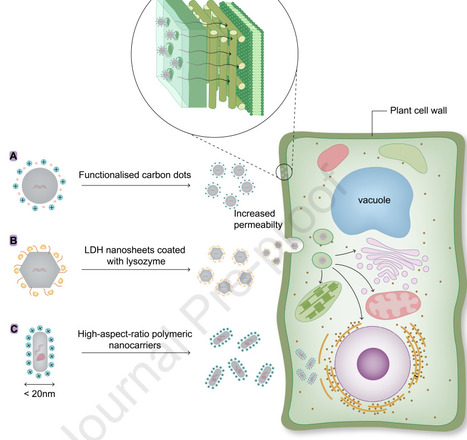
Nano-carriers are revolutionizing plant genetic engineering by facilitating the direct and efficient delivery of nucleic acids and proteins into plant cells. By precisely controlling and tailoring their properties, these carriers can effectively penetrate the plant cell wall, delivering bioactive molecules overcoming the constraints imposed by plant genotype specificity. This advancement provides a powerful, flexible, and broadly applicable strategy for accelerating crop health and resilience. Conventional plant transformation methods, such as Agrobacterium-mediated gene transfer and biolistic delivery, are limited by species-specific constraints, lengthy regeneration processes, and increased risks of genomic instability. Nanomaterial-based delivery systems offer a flexible alternative but face the significant challenge of penetrating the complex, multilayered plant cell wall. Recent research highlights three effective strategies: optimizing particle size, adjusting surface charge, and using cell wall-degrading enzymes. Together, these approaches greatly enhance nanocarrier uptake, enabling delivery of diverse bioactive agents like nucleic acids, proteins and gene-editing tools making nanotechnology a promising tool for crop improvement.

|
Scooped by
?
Today, 12:39 AM
|
Bacterial genomes exhibit significant variation in gene content and sequence identity. Pangenome analyses explore this diversity by classifying genes into core and accessory clusters of orthologous groups (COGs). However, strict sequence identity cutoffs can misclassify divergent alleles as different genes, inflating accessory gene counts. CLARC (Connected Linkage and Alignment Redefinition of COGs) (https://github.com/IndraGonz/CLARC) improves pangenome analyses by condensing accessory COGs using functional annotation and linkage information. Through this approach, orthologous groups are consolidated into more practical units of selection. Analyzing 8000+ Streptococcus pneumoniae genomes, CLARC reduced accessory gene estimates by >30% and improved evolutionary predictions based on accessory gene frequencies. CLARC is effective across different bacterial species, making it a broadly applicable tool for comparative genomics. By refining COG definitions, CLARC offers critical insights into bacterial evolution, aiding genetic studies across diverse populations.

|
Scooped by
?
Today, 12:22 AM
|
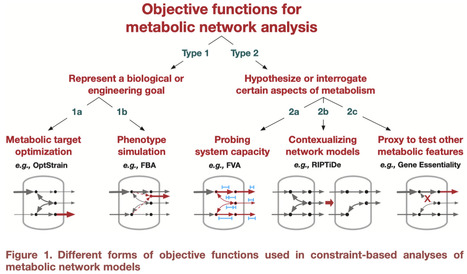
The “objective function” is a core concept in metabolic network modeling. Its use has enabled the analysis of large data to drive deeper understanding of cellular metabolism. This commentary reframes how the objective function is discussed to enhance its value and clarify misunderstandings in metabolic network modeling.

|
Scooped by
?
Today, 12:14 AM
|
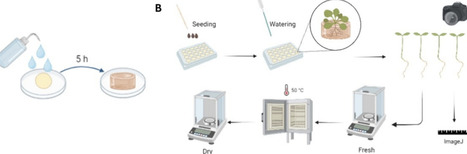
Hydrogels are emerging as sustainable alternatives to petroleum-based foams and pots in horticulture due to their high porosity, enhancing water and nutrient retention. This study develops κ-carrageenan (Carr)-based hydrogels incorporating hydrolyzed (HSW) and nonhydrolyzed red seaweed (WSW) (0–50 wt %) to introduce biostimulant properties for soilless cultivation. Hydrogels were analyzed for swelling in different media, solubility, microstructure (SEM), physicochemical interactions (FTIR, XRD), and mechanical properties, revealing high porosity, superabsorbent behavior (swelling up to 6000%), and reduced compression modulus with increasing seaweed content. Their biostimulant effect was assessed on Arabidopsis thaliana, with 20 wt % HSW hydrogels promoting enhanced root and shoot growth. Additionally, the hydrogels inhibited Fusarium solani, demonstrating antifungal properties. These results highlight the potential of Carr-seaweed hydrogels as multifunctional substrates for sustainable plant cultivation.
|

|
Scooped by
?
Today, 2:45 PM
|
The gut virome plays a pivotal role in shaping the host’s microbiota. However, gut viruses across different mammalian models, and their connections with the human gut microbiota remain largely unknown. We identified 977 high-confidence species-level viral operational taxonomic units (vOTUs) in mice (hcMGV), 12,896 in pigs (hcPGV), and 1480 in cynomolgus macaques (hcCMGV) from metagenomes, respectively. Clustering these vOTUs at approximately genus level uncovered novel clades with high prevalence across animal guts (> = 60%). In particular, crAss-like phages and cas-harboring jumbophages were characterized. Comparative analysis revealed that hcCMGV had a closer relationship with hcPGV than hcMGV, despite the animal-specific characteristics, and that 55.88% hcCMGV had connections with the human microbiota. Our findings shed light on the diversity of gut viruses across these three animals, contributing to future gut microbial studies using model animals.

|
Scooped by
?
Today, 2:39 PM
|
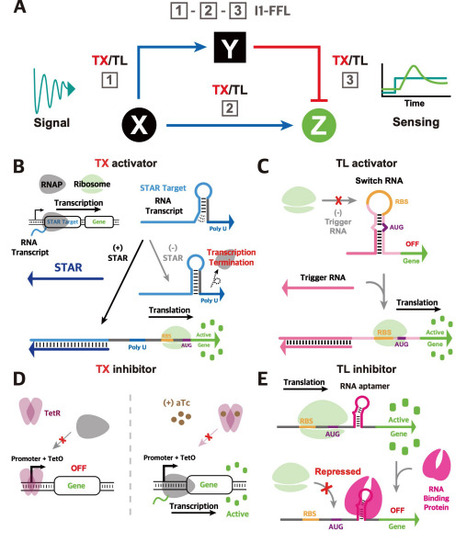
Regulating gene expression with precision is essential for cellular engineering and biosensing applications, where rapid, programmable, and sensitive control is desired. Current approaches to regulatory circuit design often rely on control at a single regulatory level, primarily the transcriptional level, thereby limiting the capability of fine-tuning the regulatory dynamics in response to complex stimuli. To address this challenge, we developed four novel RNA-protein hybrid type-1 incoherent feed-forward loop (I1-FFL) circuits in Escherichia coli that integrate transcriptional and translational regulators to achieve multi-level control of gene expression. These hybrid circuits leverage the modularity and rapid dynamics of RNA-based activators alongside the versatile inhibition capabilities of the protein-based repressors, to endow tunable pulse dynamics through engineered delays that act as transient repressor decoys. By repurposing synthetic RNA regulators at multiple regulatory levels together with aptamer and RNA-binding proteins, we demonstrate previously unexplored circuits with tunable dynamics. Complementary simulation results highlighted the importance of the engineered delays in achieving tunable pulse dynamics in these circuits. Integrating modeling insights with experimental validation, we demonstrated the flexibility of designing the RNA-protein hybrid I1-FFL circuits, as well as the tunability of their dynamics, highlighting their suitability for applications in environmental monitoring, metabolic engineering, and other engineered biological systems, where precise temporal control and adaptable gene regulation are desired.

|
Scooped by
?
Today, 2:27 PM
|
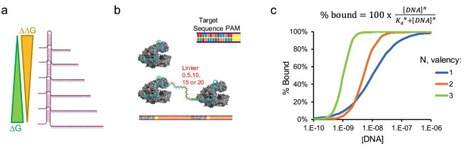
CRISPR-Cas diagnostics are revolutionizing point-of-care molecular testing due to the programmability, simplicity, and sensitivity of Cas systems with trans-cleavage activity. CRISPR-Cas12 assays are promising for detecting single nucleotide polymorphisms (SNPs). However, reports vary widely describing Cas12 SNP sensitivity, and an underlying mechanism is lacking. We systematically varied crRNA length and valency to investigate the role of crRNA architectures on Cas12 biosensing in the context of speed-of-detection, sensitivity, and selectivity. Our results demonstrate that crRNAs complementary to 20 base pairs of the target DNA is optimal for rapid and sensitive detection, while a complementary length of 15 base pairs is ideal for robust SNP detection. Additionally, we uncovered a unique periodicity in SNP sensitivity based on nucleotide position and developed a structural model explaining what drives Cas12 SNP sensitivity. Lastly, we showed that bivalent CRISPR-Cas sensors have synergistic and enhanced activity that is distance dependent. Profiling crRNAs revealed optimal length for Cas12-based molecular testing and SNP detection. Investigation into multivalent architectures elucidated valency and distance dependence for synergistic Cas12 activity.

|
Scooped by
?
Today, 2:18 PM
|
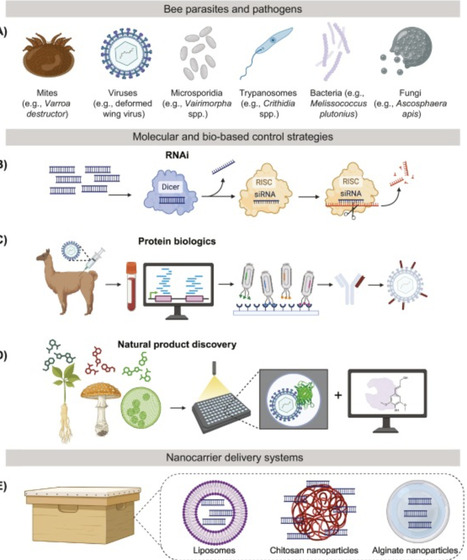
Bees are vital to global food security and biodiversity but their populations are threatened by a steady flux of interacting stressors. Current mitigation strategies are failing to address the complexity and scale of these threats. Biotechnology offers innovative solutions to protect essential pollination services and secure the future of beekeeping. Omic tools guided by artificial intelligence can unlock new possibilities for strengthening bee populations and improve their ability to adapt to emerging challenges. Molecular and bio-based treatments offer precise, nonchemical inputs for managed hives. Synthetic biology enables engineered gut microbiomes, pollinator-friendly crops, and artificial diets that are tailored to bee health. We discuss recent progress and future directions of biotechnology to help bees cope with a rapidly changing world.

|
Scooped by
?
Today, 1:31 PM
|
Cyanidiophyceae red algae dominate many geothermal habitats and provide important tools for investigating the evolution of extremophilic eukaryotes and associated microbial communities. We propose that resource sharing drove genome reduction in Cyanidiophyceae and enabled the neofunctionalization of genes in multi-enzyme pathways. Utilizing arsenic detoxification as a model, we discuss how the sharing of gene functions by other members of the microbial assemblage weakened selection on homologs in the Cyanidiophyceae, allowing long-term gene persistence via the putative gain of novel functions. This hypothesis, referred to as the Integrated Horizontal Gene Transfer (HGT) Model (IHM), attempts more generally to explain how extremophilic eukaryotes may have transitioned from 'hot start' milieus by functional innovations driven by the duplication and divergence of HGT-derived genes.

|
Scooped by
?
Today, 1:26 PM
|
Big Science projects are often troubled by misalignments. We analyze how synthetic cell researchers handle misalignments by performing alignment work. We find that alignment work renders Big Science feasible, but involves trade-offs, for example between short-term successes and long-term goals. We thus recommend attending to the politics of alignment work.

|
Scooped by
?
Today, 1:13 PM
|
Traditionally perceived as an RNA-specific nuclease, Cas13a has been used primarily for RNA detection. We discover the ability of Leptotrichia buccalis Cas13a (LbuCas13a) to directly target DNA without the restrictions of protospacer flanking sequence and PAM sequences, coupled with robust trans-cleavage activity. Contrary to conventional understanding, LbuCas13a does not degrade DNA targets. Our study reveals an enhancement in the single-nucleotide specificity of LbuCas13a against DNA compared to RNA. This heightened specificity is attributed to the lower affinity of CRISPR RNA (crRNA) towards DNA, raising the crRNA–DNA binding energy barrier. We introduce a molecular diagnostic platform called superior universal rapid enhanced specificity test with LbuCas13a (SUREST) for high-resolution genotyping. SUREST is capable of detecting DNA concentrations of CYP2C19 (rs4986893) as minute as 0.3 aM (0.18 cps µl−1). We also apply SUREST to human genotyping scenarios, indicating that SUREST performs well across a broad range of mutations and sequence contexts. SUREST represents an advancement in real-time nucleic acid detection, making it a useful tool for pathogen identification and mutation analysis in clinical diagnostics. A Leptotrichia buccalis Cas13a diagnostic platform can be used to cleave and detect small concentrations of viral and human DNA.

|
Scooped by
?
Today, 1:00 PM
|
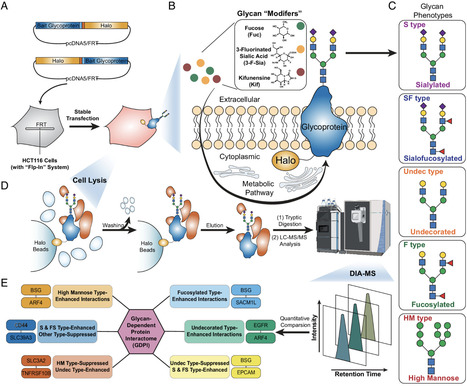
Protein–protein interactions (PPI) are crucial for comprehending the molecular mechanisms and signaling pathways underlying diverse biological processes and disease progression. However, investigating PPIs involving membrane proteins is challenging due to the complexity and heterogeneity of glycosylation. To tackle this challenge, we developed an approach termed glycan-dependent affinity purification coupled with mass spectrometry (GAP–MS), specifically designed to characterize changes in glycoprotein PPIs under varying glycosylation conditions. GAP–MS integrates metabolic control of glycan profiles in cultured cells using small molecules referred to as glycan modifiers with affinity purification followed by mass spectrometry analysis (AP–MS). Here, GAP–MS was applied to characterize and compare the interaction networks under five different glycosylation states for four bait glycoproteins: BSG, CD44, EGFR, and SLC3A2. This analysis identified a network comprising 156 interactions, of which 131 were determined to be glycan dependent. Notably, the GAP–MS analysis of BSG provided distinct information regarding glycosylation-influenced interactions compared to the commonly used glycosylation site mutagenesis approach combined with AP–MS, emphasizing the unique advantages of GAP–MS. Collectively, GAP–MS presents distinct insights over existing methods in elucidating how specific glycosylation forms impact glycoprotein interactions. Additionally, the glycan-dependent interaction networks generated for these four glycoproteins serve as a valuable resource for guiding future functional investigations and therapeutic developments targeting the glycoproteins discussed in this study.

|
Scooped by
?
Today, 1:04 AM
|
Testing microbial interaction hypotheses is a major challenge in microbiome research. Bacteriophages may be used to probe organism-organism interactions within complex microbiomes, yet are rarely used for this purpose. Here, we isolated nine narrow host range phages that replicate in several Variovorax species. As Variovorax CL14 degrades the plant root stunting hormone auxin, we used phages to eliminate V. CL14 from a rhizosphere consortium and reestablished the stunted root phenotype. We used three of the phages to deplete another Variovorax, SCN45, from complex communities to test correlation network-based hypotheses of thiamine interdependencies. Genome-resolved metagenomics revealed that three taxa decreased in relative abundance following Variovorax depletion and could be rescued by thiamine supplementation. Thus, we confirmed thiamine production as the mechanistic basis for interdependence. These experiments lay the foundation for research that employs wildtype and genetically modified phages to test interaction hypotheses and for targeted microbiome manipulation.

|
Scooped by
?
Today, 12:54 AM
|
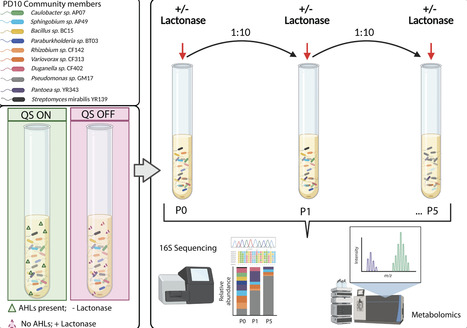
Bacteria are recognized for their diverse metabolic capabilities, yet the impact of microbe-microbe interactions on multispecies community structure and dynamics is poorly understood. Cell-to-cell signaling in the form of quorum sensing (QS) often regulates secondary metabolite production and microbial interactions. Here, we examine how acylhomoserine lactone (AHL)-mediated QS impacts microbial community structure in a 10-member synthetic community of isolates from Populus deltoides. To explore the role of QS in microbial community structure and dynamics, we disrupted AHL signaling by exogenous addition of AiiA-lactonase, an enzyme that cleaves the lactone ring. Microbial community structure resulting from signal inactivation, as measured by 16S rRNA amplicon sequencing and secondary metabolite production, was assessed after successive passaging of the community. Further, we investigated the impact of quorum quenching on specific microbe-microbe interactions using pairwise inhibition assays. Our results indicate that AHL inactivation alters the relative abundance of dominant community members at later passages but does not impact the overall membership in the community. Quorum quenching significantly alters the metabolic profile in lactonase-treated communities. This metabolic alteration impacts microbe-microbe interactions through decreased inhibition of other community members. Together, these results indicate that QS impacts microbial community structure through the regulation of secondary metabolites in dominant members and that the membership of microbial communities can be relatively stable despite changes in metabolic profiles.

|
Scooped by
?
Today, 12:32 AM
|
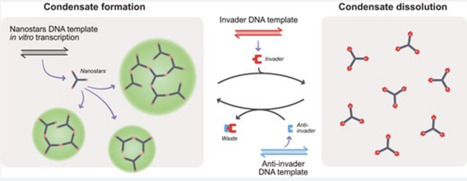
RNA-driven phase separation is emerging as a promising approach for engineering biomolecular condensates with diverse functionalities. Condensates form thanks to weak yet specific RNA–RNA interactions established by design via complementary sequence domains. Here, we demonstrate how RNA condensates formed by star-shaped RNA motifs, or nanostars, can be dynamically controlled when the motifs include additional linear or branch-loop domains that facilitate access of regulatory RNA molecules to the nanostar interaction domains. We show that condensates dissolve in the presence of RNA “invaders” that occlude selected nanostar bonds and reduce the valency of the nanostars, preventing phase separation. We further demonstrate that the introduction of “anti-invader” strands, complementary to the invaders, makes it possible to restore condensate formation. An important aspect of our experiments is that we demonstrate these behaviors in one-pot reactions, where RNA nanostars, invaders, and anti-invaders are simultaneously transcribed in vitro using short DNA templates. Our results lay the groundwork for engineering RNA-based assemblies with tunable, reversible condensation, providing a promising toolkit for synthetic biology applications requiring responsive, self-organizing biomolecular materials.

|
Scooped by
?
Today, 12:17 AM
|
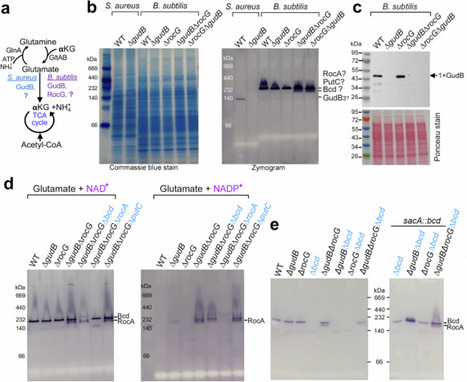
Glutamate dehydrogenase (GDH) resides at the crossroads of nitrogen and carbon metabolism, catalyzing the reversible conversion of L-glutamate to α-ketoglutarate and ammonium. GDH paralogs are ubiquitous across most species, presumably enabling functional specialization and genetic compensation in response to diverse conditions. Staphylococcus aureus harbors a single housekeeping GDH (GudB), whereas Bacillus subtilis encodes both a major and a minor GDH, GudB and RocG, respectively. In an unsuccessful attempt to identify an alternative GDH in S. aureus, we serendipitously discovered previously unrecognized GDH activity in two metabolic enzymes of B. subtilis. The hexameric Val/Leu/Ile dehydrogenase Bcd (formerly YqiT) catabolizes branched-chain amino acids and to a lesser extent glutamate using NAD+ as a cofactor. Removal of gudB and rocG unmasks the dual NAD(P)+-dependent GDH activity of RocA, which otherwise functions as a 3-hydroxy-1-pyrroline-5-carboxylate dehydrogenase. Bcd homologs are prevalent in free-living and obligate bacteria but are absent in most, if not all, staphylococci. Despite low sequence homology, Bcd structurally resembles the GudB/RocG family and can functionally compensate for the loss of GudB in S. aureus. Bcd is essential for the full maturation of biofilms. B. subtilis lacking GDHs exhibits severe impairments in rugose architecture and colony expansion of biofilms. This study underscores the importance of metabolic redundancy and highlights the critical role of substrate promiscuity in GDHs during biofilm development.
|
 Your new post is loading...
Your new post is loading...














omv