Your new post is loading...
 Your new post is loading...

|
Scooped by
The Sainsbury Lab
April 28, 3:41 PM
|
Key message Plant U-box E3 ligases PUB20 and PUB21 are flg22-triggered signaling components and negatively regulate immune responses. Abstract Plant U-box proteins (PUBs) constitute a class of E3 ligases that are associated with various stress responses. Among the class IV PUBs featuring C-terminal Armadillo (ARM) repeats, PUB20 and PUB21 are closely related homologs. Here, we show that both PUB20 and PUB21 negatively regulate innate immunity in plants. Loss of PUB20 and PUB21 function leads to enhanced resistance to surface inoculation with the virulent bacterium Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). However, the resistance levels remain unaffected after infiltration inoculation, suggesting that PUB20 and PUB21 primarily function during the early defense stages. The enhanced resistance to Pst DC3000 in PUB mutant plants (pub20-1, pub21-1, and pub20-1/pub21-1) correlates with extensive flg22-triggered reactive oxygen production, strong MPK3 activation, and enhanced transcriptional activation of early immune response genes. Additionally, PUB mutant plants (except pub21-1) exhibit constitutive stomatal closure after Pst DC3000 inoculation, implying the significant role of PUB20 in stomatal immunity. Comparative analyses of flg22 responses between PUB mutants and wild-type plants reveals that the robust activation of the pattern-induced immune responses may enhance resistance against Pst DC3000. Notably, the hypersensitivity responses triggered by RPM1/avrRpm1 and RPS2/avrRpt2 are independent of PUB20 and PUB21. These results suggest that PUB20 and PUB21 knockout mutations affect bacterial invasion, likely during the early stages, acting as negative regulators of plant immunity.

|
Scooped by
The Sainsbury Lab
April 28, 3:12 PM
|
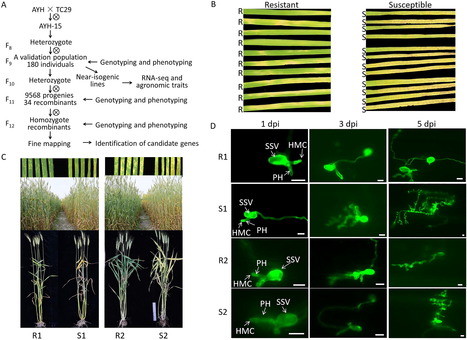
Stripe rust, caused by Puccinia striiformis f. sp. tritici ( Pst), is a devastating disease in wheat worldwide. Discovering and characterizing new resistance genes/QTL is crucial for wheat breeding programs. In this study, we fine-mapped and characterized a stripe rust resistance gene, YRAYH, on chromosome arm 5BL in the Chinese wheat landrace Anyuehong (AYH). Evaluations of stripe rust response to prevalent Chinese Pst races in near-isogenic lines derived from a cross of Anyuehong and Taichung 29 showed that YrAYH conferred a high level of resistance at all growth stages. Fine mapping using a large segregating population of 9748 plants, narrowed the YRAYH locus to a 3.7 Mb interval on chromosome arm 5BL that included 61 annotated genes. Transcriptome analysis of two NIL pairs identified 64 upregulated differentially expressed genes (DEGs) in the resistant NILs (NILs-R). Annotations indicated that many of these genes have roles in plant disease resistance pathways. Through a combined approach of fine-mapping and transcriptome sequencing, we identified a serine/threonine-protein kinase SRPK as a candidate gene underlying YrAYH. A unique 25 bp insertion was identified in the NILs-R compared to the NILs-S and previously published wheat genomes. An InDel marker was developed and co-segregated with YrAYH. Agronomic trait evaluation of the NILs suggested that YrAYH not only reduces the impact of stripe rust but was also associated with a gene that increases plant height and spike length.

|
Scooped by
The Sainsbury Lab
February 12, 5:01 AM
|
-
Asian soybean rust (ASR), caused by the obligate biotrophic fungus, Phakopsora pachyrhizi, was first reported in the continental United States of America (USA) in 2004 and over the years has been of concern to soybean production in the USA. The prevailing hypothesis is that P. pachyrhizi spores were introduced into the USA via hurricanes originating from South America, particularly Hurricane Ivan. -
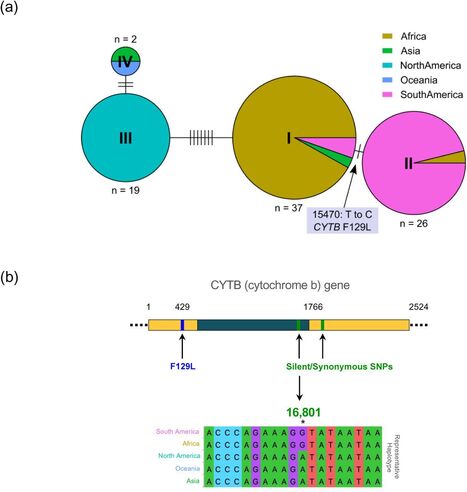
To investigate the genetic diversity and global population structure of P. pachyrhizi, we employed exome-capture based sequencing on 84 field isolates collected from different geographic regions worldwide. We compared the gene-encoding regions from all these field isolates and found that four major haplotypes are prevalent worldwide. Here, we provide genetic evidence supporting multiple incursions that have led to the currently established P. pachyrhizi population of the USA. Phylogenetic analysis of mitochondrial genes further supports this hypothesis. -
Notably, we observed limited genetic diversity in P. pachyrhizi populations in Brazil, suggesting a clonal population structure in that country that contrasts to populations from the USA and Africa. -
This study provides the first comprehensive characterization of P. pachyrhizi population structures defined by genetic evidence from populations across major soybean growing regions.

|
Scooped by
The Sainsbury Lab
February 12, 4:43 AM
|
Carbohydrate-based cell wall signaling impacts plant growth, development, and stress responses; however, how cell wall signals are perceived and transduced remains poorly understood. Several cell wall breakdown products have been described as typical damage-associated molecular patterns that activate plant immunity, including pectin-derived oligogalacturonides (OGs). Receptor kinases of the WALL-ASSOCIATED KINASE (WAK) family bind pectin and OGs and were previously proposed as OG receptors. However, unambiguous genetic evidence for the role of WAKs in OG responses is lacking. Here, we investigated the role of Arabidopsis (Arabidopsis thaliana) WAKs in OG perception using a clustered regularly interspaced short palindromic repeats mutant in which all 5 WAK genes were deleted. Using a combination of immune assays for early and late pattern-triggered immunity, we show that WAKs are dispensable for OG-induced signaling and immunity, indicating that they are not bona fide OG receptors.

|
Scooped by
The Sainsbury Lab
February 12, 4:40 AM
|
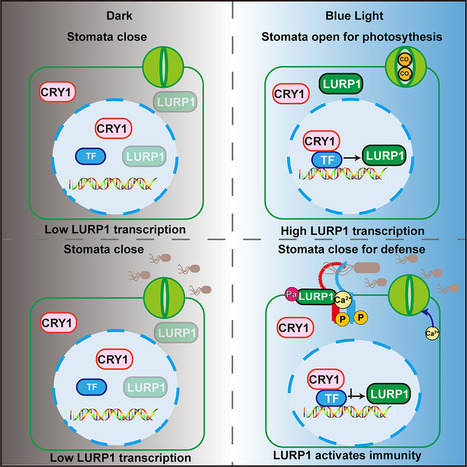
Plant stomata open in response to blue light, allowing gas exchange and water transpiration. However, open stomata are potential entry points for pathogens. Whether plants can sense pathogens and mount defense responses upon stomatal opening and how blue-light cues are integrated to balance growth-defense trade-offs are poorly characterized. We show that the Arabidopsis blue-light photoreceptor CRYPTOCHROME 1 (CRY1) mediates various aspects of immunity, including pathogen-triggered stomatal closure as well as activation of plant immunity through a typical light-responsive protein LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA (LURP1). LURP1 undergoes N-terminal palmitoylation in the presence of bacterial flagellin, prompting a change in subcellular localization from the cytoplasm to plasma membrane, where it enhances the activity of the receptor FLAGELLIN SENSING 2 (FLS2) to mediate plant defense. Collectively, these findings reveal that blue light regulates stomatal defense and highlight the dual functions of CRY1 in photosynthesis and immunity.

|
Scooped by
The Sainsbury Lab
February 12, 4:31 AM
|
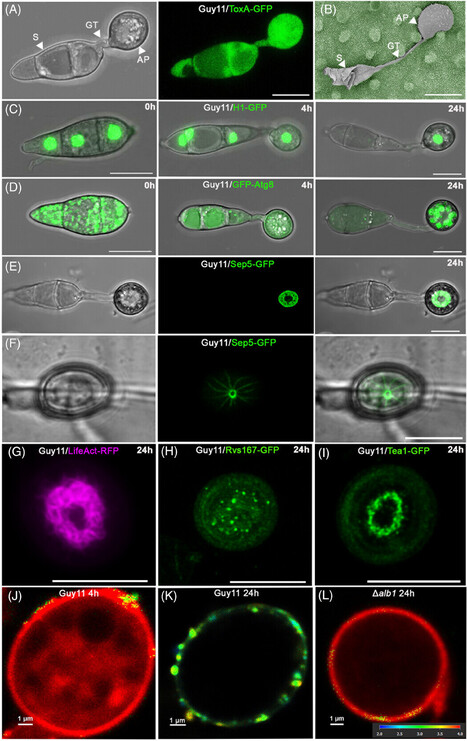
Magnaporthe oryzae is the causal agent of rice blast, one of the most serious diseases affecting rice cultivation around the world. During plant infection, M. oryzae forms a specialised infection structure called an appressorium. The appressorium forms in response to the hydrophobic leaf surface and relies on multiple signalling pathways, including a MAP kinase phosphorelay and cAMP-dependent signalling, integrated with cell cycle control and autophagic cell death of the conidium. Together, these pathways regulate appressorium morphogenesis.The appressorium generates enormous turgor, applied as mechanical force to breach the rice cuticle. Re-polarisation of the appressorium requires a turgor-dependent sensor kinase which senses when a critical threshold of turgor has been reached to initiate septin-dependent re-polarisation of the appressorium and plant infection. Invasive growth then requires differential expression and secretion of a large repertoire of effector proteins secreted by distinct secretory pathways depending on their destination, which is also governed by codon usage and tRNA thiolation. Cytoplasmic effectors require an unconventional Golgi-independent secretory pathway and evidence suggests that clathrin-mediated endocytosis is necessary for their delivery into plant cells. The blast fungus then develops a transpressorium, a specific invasion structure used to move from cell-to-cell using pit field sites containing plasmodesmata, to facilitate its spread in plant tissue. This is controlled by the same MAP kinase signalling pathway as appressorium development and requires septin-dependent hyphal constriction. Recent progress in understanding the mechanisms of rice infection by this devastating pathogen using live cell imaging procedures are presented.

|
Scooped by
The Sainsbury Lab
February 11, 3:44 PM
|
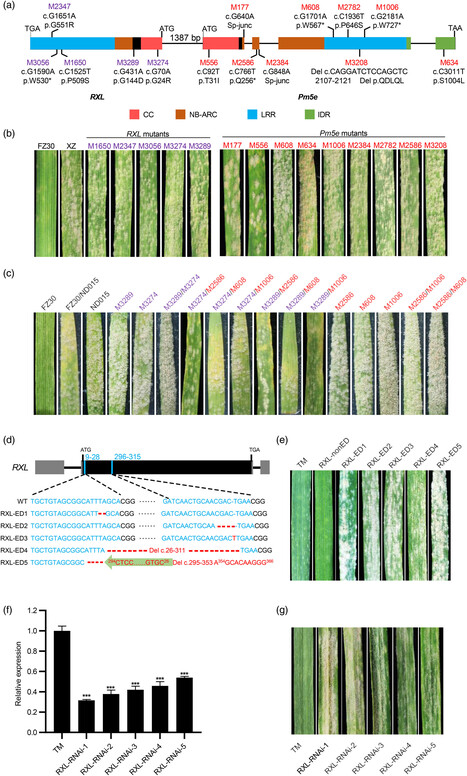
Powdery mildew poses a significant threat to global wheat production and most cloned and deployed resistance genes for wheat breeding encode nucleotide-binding and leucine-rich repeat (NLR) immune receptors. Although two genetically linked NLRs function together as an NLR pair have been reported in other species, this phenomenon has been relatively less studied in wheat. Here, we demonstrate that two tightly linked NLR genes, RXL and Pm5e, arranged in a head-to-head orientation, function together as an NLR pair to mediate powdery mildew resistance in wheat. The resistance function of the RXL/Pm5e pair is validated by mutagenesis, gene silencing, and gene-editing assays. Interestingly, both RXL and Pm5e encode atypical NLRs, with RXL possessing a truncated NB-ARC (nucleotide binding adaptor shared by APAF-1, plant R proteins and CED-4) domain and Pm5e featuring an atypical coiled-coil (CC) domain. Notably, RXL and Pm5e lack an integrated domain associated with effector recognition found in all previously reported NLR pairs. Additionally, RXL and Pm5e exhibit a preference for forming hetero-complexes rather than homo-complexes, highlighting their cooperative role in disease resistance. We further show that the CC domain of Pm5e specifically suppresses the hypersensitive response induced by the CC domain of RXL through competitive interaction, revealing regulatory mechanisms within this NLR pair. Our study sheds light on the molecular mechanism underlying RXL/Pm5e-mediated powdery mildew resistance and provides a new example of an NLR pair in wheat disease resistance.

|
Scooped by
The Sainsbury Lab
February 11, 3:24 PM
|
Soybean [Glycine max (L.) Merrill] is one of the most widely grown legumes in the world, with Brazil being its largest producer and exporter. Breeding programs in Brazil have resulted from multiple cycles of selection and recombination starting from a small number of USA cultivar ancestors in the 1950s and 1960s years. This process has led to the successful adaptation of this crop to tropical conditions, a phenomenon known as tropicalization. Many studies describe a narrow genetic background in Brazilian soybean cultivars. Various factors can affect the genetic diversity in species, especially in cultivated crops, such as the reproduction type, artificial selection, and the number and sources of variability in the breeding programs. In turns, the genetic diversity can affect the linkage disequilibrium blocks (LD) patterns and, consequently, molecular breeding strategies for selection of target loci for agronomic traits. We used high-throughput genotyping with SoySNP50K Illumina SNP markers to assess a collection of 370 Brazilian soybean accessions covering more than 60 years of soybean breeding in Brazil. Our goal was to investigate population structure and genetic diversity in the Brazilian germplasm, detect patterns of LD blocks, and identify regions presenting signals of selective swaps linked with quantitative trait loci (QTLs) of agronomic interest. Population structure analysis revealed two major groups among all genotypes, primarily differentiated by the year of release, separating old and new cultivars (before and after 2000´s years), and by growth habit (stem termination type—SST). The group I comprises about 75% of the panel and includes cultivars release before 2000`s years, including the oldest cultivars released in Brazil, most of which exhibit a determinate growth habit and maturity groups VI and VII. Group II includes only 83 materials, but shows higher levels of diversity than group I, representing most recent introductions in Brazilian germplasm. Further analysis of substructure within Group I, identified seven subgroups with no clear trend for segregation based on maturity group, STT or year of release. Instead, these subgroups were based on the contribution of key donors of disease resistance and adaptability, as soybean cultivation expanded from the South to Central region of Brazil. This finding is consistent with the history of soybean expansion in Brazil. We identified 123 genomic regions under selection among the groups of Brazilian cultivars associated with 440 quantitative trait loci (QTLs), revealing regions fixed across the breeding process associated with yield, disease resistance, water efficiency use, and others.

|
Scooped by
The Sainsbury Lab
December 12, 2024 12:01 PM
|
Plants have evolved an elaborate cell wall integrity (CWI) sensing system to monitor and modify cell wall formation. LRR-extensins (LRXs) are cell wall-anchored proteins that bind RAPID ALKALINIZATION FACTOR (RALF) peptide hormones and induce compaction of cell wall structures. At the same time, LRXs form a signaling platform with RALFs and the transmembrane receptor kinase FERONIA (FER) as a means to relay changes in CWI to the protoplast. LRX1 of Arabidopsis thaliana is predominantly expressed in root hairs and lrx1mutants develop defective root hairs. Here, we identify a regulator of LRX1-RALF-FER signaling as a suppressor of the lrx1 root hair phenotype. The repressor of lrx1_23 (rol23) gene encodes PP2C12, a type 2C phosphatase of clade H that interacts with FER and dephosphorylates Thr696 in the FER activation loop in vitro. The LRX1-related function of PP2Cs appears clade H-specific and was not observed for other PP2Cs investigated. Collectively, our data suggest that LRX1 acts upstream of the RALF1-FER signaling module and PP2C12 has an inhibitory activity via modulating FER activity to fine-tune CWI signaling.

|
Scooped by
The Sainsbury Lab
December 12, 2024 11:38 AM
|
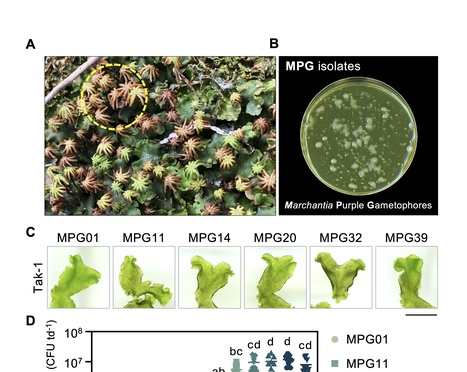
Plant pathogenic Pseudomonas species naturally antagonize a diverse range of flowering plants. While emerging research demonstrates that isolates belonging to the P. syringaespecies complex colonize diverse hosts, the extent to which these bacteria naturally infect non-flowering plants like the model liverwort Marchantia polymorpha remains unclear. Here, we identify natural associations between Pseudomonas viridiflava and the liverwort Marchantia polymorpha. Pseudomonas bacteria isolated from diseased liverworts in the wild successfully re-infected M. polymorpha in pure culture conditions, producing high in planta bacterial densities and causing prominent tissue maceration. Comparative genomic analysis of Marchantia-associated P. viridiflava identified core virulence machinery like the type-III secretion system (T3SS) and conserved effectors (AvrE and HopM1) that were essential for liverwort infection. Disease assays performed in Nicotiana benthamiana further confirmed that liverwort-associated P. viridiflava infect flowering plants in an effector-dependent manner. Our work highlights P. viridiflava as an effective broad host pathogen that relies on conserved virulence factors to manipulate evolutionarily divergent host plants.

|
Scooped by
The Sainsbury Lab
December 12, 2024 11:15 AM
|
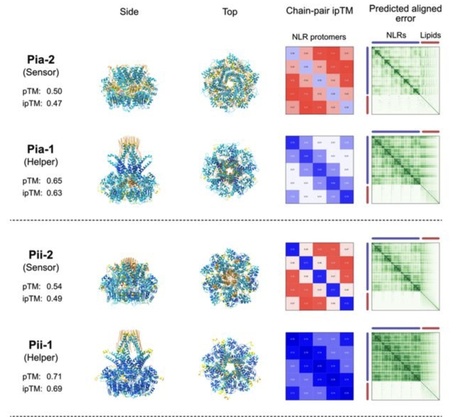
NLR immune receptors can be functionally organized in genetically linked sensor-helper pairs. However, methods to categorize paired NLRs remain limited, primarily relying on the presence of non-canonical domains in some sensor NLRs. Here, we propose that the AI system AlphaFold 3 can classify paired NLR proteins into sensor or helper categories based on predicted structural characteristics. Helper NLRs showed higher AlphaFold 3 confidence scores than sensors when modelled in oligomeric configurations. Furthermore, funnel-shaped structures—essential for activating immune responses—were reliably predicted in helpers but not in sensors. Applying this method to uncharacterized NLR pairs from rice, we found that AlphaFold 3 can differentiate between putative sensors and helpers even when both proteins lack non-canonical domain annotations. These findings suggest that AlphaFold 3 offers a new approach to categorize NLRs and enhances our understanding of the functional configurations in plant immune systems, even in the absence of non-canonical domain annotations.

|
Scooped by
The Sainsbury Lab
November 18, 2024 5:22 PM
|
Nucleotide-binding leucine-rich repeat (NLR) disease resistance genes typically confer resistance against races of a single pathogen. Here, we report that Yr87/Lr85, an NLR gene from Aegilops sharonensis and Aegilops longissima, confers resistance against both P. striiformis tritici (Pst) and Puccinia triticina (Pt) that cause stripe and leaf rust, respectively. Yr87/Lr85 confers resistance against Pst and Pt in wheat introgression as well as transgenic lines. Comparative analysis of Yr87/Lr85 and the cloned Triticeae NLR disease resistance genes shows that Yr87/Lr85 contains two distinct LRR domains and that the gene is only found in Ae. sharonensis and Ae. longissima. Allele mining and phylogenetic analysis indicate multiple events of Yr87/Lr85 gene flow between the two species and presence/absence variation explaining the majority of resistance to wheat leaf rust in both species. The confinement of Yr87/Lr85 to Ae. sharonensis and Ae. longissima and the resistance in wheat against Pst and Pt highlight the potential of these species as valuable sources of disease resistance genes for wheat improvement. Leaf rust and stripe rust of wheat are two important fungal diseases of cultivated wheat and they are caused by infection of different pathogens. Here, the authors report the nucleotide-binding leucine-rich repeat (NLR) protein encoding gene Yr87/Lr85 confers resistance to both diseases.

|
Scooped by
The Sainsbury Lab
November 18, 2024 5:19 PM
|
The plant immune system relies on germline-encoded pattern recognition receptors (PRRs) that sense foreign and plant-derived molecular patterns, and signal health threats. Genomic and pangenomic data sets provide valuable insights into the evolution of PRRs and their molecular triggers, which is furthering our understanding of plant–pathogen co-evolution and convergent evolution. Moreover, in silico and in vivo methods of PRR identification have accelerated the characterization of receptor–ligand complexes, and advances in protein structure prediction algorithms are revealing novel PRR sensor functions. Harnessing these recent advances to engineer PRRs presents an opportunity to enhance plant disease resistance against a broad spectrum of pathogens, enabling more sustainable agricultural practices. This Review summarizes both established and innovative approaches to leverage genomic data and translate resulting evolutionary insights into engineering PRR recognition specificities. Genomic and pangenomic data are yielding insights into the evolution of plant pattern recognition receptors (PRRs) and their molecular triggers. Recent advances in in silico and in vivo methods, alongside protein structure prediction, are helping to harness these insights for PRR engineering, offering sustainable solutions for broad-spectrum plant disease resistance.
|

|
Scooped by
The Sainsbury Lab
April 28, 3:26 PM
|
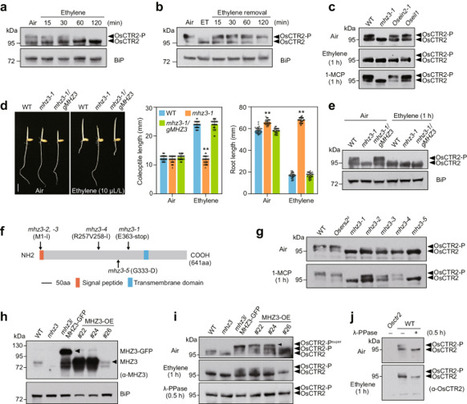
Ethylene regulates plant growth, development, and stress adaptation. However, the early signaling events following ethylene perception, particularly in the regulation of ethylene receptor/CTRs (CONSTITUTIVE TRIPLE RESPONSE) complex, remains less understood. Here, utilizing the rapid phospho-shift of rice OsCTR2 in response to ethylene as a sensitive readout for signal activation, we revealed that MHZ3, previously identified as a stabilizer of ETHYLENE INSENSITIVE 2 (OsEIN2), is crucial for maintaining OsCTR2 phosphorylation. Genetically, both functional MHZ3 and ethylene receptors prove essential for OsCTR2 phosphorylation. MHZ3 physically interacts with both subfamily I and II ethylene receptors, e.g., OsERS2 and OsETR2 respectively, stabilizing their association with OsCTR2 and thereby maintaining OsCTR2 activity. Ethylene treatment disrupts the interactions within the protein complex MHZ3/receptors/OsCTR2, reducing OsCTR2 phosphorylation and initiating downstream signaling. Our study unveils the dual role of MHZ3 in fine-tuning ethylene signaling activation, providing insights into the initial stages of the ethylene signaling cascade. The early signalling events following ethylene perception by plants remain incompletely understood. Here the authors show that in the absence of ethylene, rice MHZ3, a known stabilizer of OsEIN2, promotes phosphorylation of OsCTR2 to suppress ethylene signalling.

|
Scooped by
The Sainsbury Lab
February 12, 5:09 AM
|
-
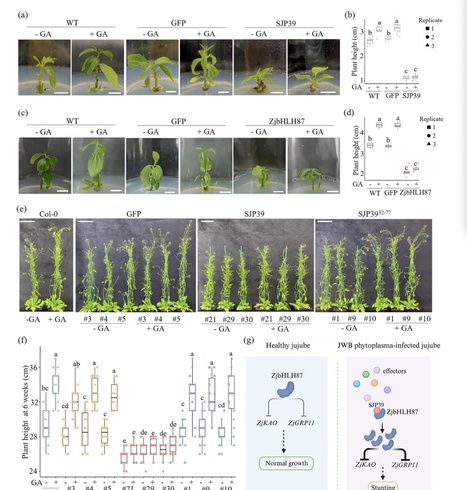
Phytoplasmas are specialized phloem-limited bacteria that cause diseases on various crops resulting in significant agricultural losses. This research focuses on the jujube Witches’ Broom (JWB) phytoplasma and investigates the host-manipulating activity of the effector SJP39. -
We found that SJP39 directly interacts with the plant transcription factor bHLH87 in the nuclei. SJP39 stabilizes the bHLH87 homologs in A. thaliana and jujube, leading to growth defects in the plants. -
Transcriptomic analysis indicates that SJP39 affects the gibberellin (GA) pathway in jujube. We further demonstrate that ZjbHLH87 regulates GA signalling as a negative regulator and SJP39 enhances this regulation. -
The research offers important insights into the pathogenesis of JWB disease and identified SJP39 as a virulence factor that can contribute to the growth defects caused by JWB phytoplasma infection. These findings open new opportunities to manage JWB and other phytoplasma diseases.

|
Scooped by
The Sainsbury Lab
February 12, 4:48 AM
|
The ability of plants to perceive and react to biotic and abiotic stresses is critical for their health. We recently identified a core set of genes consistently induced by members of the leaf microbiota, termed general non-self response (GNSR) genes. Here we show that GNSR components conversely impact leaf microbiota composition. Specific strains that benefited from this altered assembly triggered strong plant responses, suggesting that the GNSR is a dynamic system that modulates colonization by certain strains. Examination of the GNSR to live and inactivated bacteria revealed that bacterial abundance, cellular composition and exposure time collectively determine the extent of the host response. We link the GNSR to pattern-triggered immunity, as diverse microbe- or danger-associated molecular patterns cause dynamic GNSR gene expression. Our findings suggest that the GNSR is the result of a dose-responsive perception and signalling system that feeds back to the leaf microbiota and contributes to the intricate balance of plant–microbiome interactions. The plant general non-self response system is triggered by leaf microbiota members and, in turn, impacts their colonization.

|
Scooped by
The Sainsbury Lab
February 12, 4:41 AM
|
Plants lack specialized and mobile immune cells. Consequently, any cell type that encounters pathogens must mount immune responses and communicate with surrounding cells for successful defence. However, the diversity, spatial organization and function of cellular immune states in pathogen-infected plants are poorly understood1. Here we infect Arabidopsis thaliana leaves with bacterial pathogens that trigger or supress immune responses and integrate time-resolved single-cell transcriptomic, epigenomic and spatial transcriptomic data to identify cell states. We describe cell-state-specific gene-regulatory logic that involves transcription factors, putative cis-regulatory elements and target genes associated with disease and immunity. We show that a rare cell population emerges at the nexus of immune-active hotspots, which we designate as primary immune responder (PRIMER) cells. PRIMER cells have non-canonical immune signatures, exemplified by the expression and genome accessibility of a previously uncharacterized transcription factor, GT-3A, which contributes to plant immunity against bacterial pathogens. PRIMER cells are surrounded by another cell state (bystander) that activates genes for long-distance cell-to-cell immune signalling. Together, our findings suggest that interactions between these cell states propagate immune responses across the leaf. Our molecularly defined single-cell spatiotemporal atlas provides functional and regulatory insights into immune cell states in plants. The development of a molecularly defined spatiotemporal atlas of pathogen-infected Arabidopsis thaliana leaves reveals specific cell states that have distinct roles in plant immunity.

|
Scooped by
The Sainsbury Lab
February 12, 4:36 AM
|
Fungi are the most important group of plant pathogens, responsible for many of the world’s most devastating crop diseases. One of the reasons they are such successful pathogens is because several fungi have evolved the capacity to breach the tough outer cuticle of plants using specialized infection structures called appressoria. This is exemplified by the filamentous ascomycete fungus Magnaporthe oryzae, causal agent of rice blast, one of the most serious diseases affecting rice cultivation globally. M. oryzae develops a pressurized dome-shaped appressorium that uses mechanical force to rupture the rice leaf cuticle. Appressoria form in response to the hydrophobic leaf surface, which requires the Pmk1 MAP kinase signalling pathway, coupled to a series of cell-cycle checkpoints that are necessary for regulated cell death of the fungal conidium and development of a functionally competent appressorium. Conidial cell death requires autophagy, which occurs within each cell of the spore, and is regulated by components of the cargo-independent autophagy pathway. This results in trafficking of the contents of all three cells to the incipient appressorium, which develops enormous turgor of up to 8.0 MPa, due to glycerol accumulation, and differentiates a thickened, melanin-lined cell wall. The appressorium then re-polarizes, re-orienting the actin and microtubule cytoskeleton to enable development of a penetration peg in a perpendicular orientation, that ruptures the leaf surface using mechanical force. Re-polarization requires septin GTPases which form a ring structure at the base of the appressorium, which delineates the point of plant infection, and acts as a scaffold for actin re-localization, enhances cortical rigidity, and forms a lateral diffusion barrier to focus polarity determinants that regulate penetration peg formation. Here we review the mechanism of regulated cell death in M. oryzae, which requires autophagy but may also involve ferroptosis. We critically evaluate the role of regulated cell death in appressorium morphogenesis and examine how it is initiated and regulated, both temporally and spatially, during plant infection. We then use this synopsis to present a testable model for control of regulated cell death during appressorium-dependent plant infection by the blast fungus.

|
Scooped by
The Sainsbury Lab
February 11, 3:46 PM
|
Nutrient acquisition is crucial for sustaining life. Plants develop beneficial intracellular partnerships with arbuscular mycorrhiza (AM) and nitrogen-fixing bacteria to surmount the scarcity of soil nutrients and tap into atmospheric dinitrogen, respectively1,2. Initiation of these root endosymbioses requires symbiont-induced oscillations in nuclear calcium (Ca2+) concentrations in root cells3. How the nuclear-localized ion channels, cyclic nucleotide-gated channel (CNGC) 15 and DOESN’T MAKE INFECTIONS1 (DMI1)4 are coordinated to specify symbiotic-induced nuclear Ca2+ oscillations remains unknown. Here we discovered an autoactive CNGC15 mutant that generates spontaneous low-frequency Ca2+ oscillations. While CNGC15 produces nuclear Ca2+ oscillations via a gating mechanism involving its helix 1, DMI1 acts as a pacemaker to specify the frequency of the oscillations. We demonstrate that the specificity of symbiotic-induced nuclear Ca2+ oscillations is encoded in its frequency. A high frequency activates endosymbiosis programmes, whereas a low frequency modulates phenylpropanoid pathways. Consequently, the autoactive cngc15 mutant, which is capable of generating both frequencies, has increased flavonoids that enhance AM, root nodule symbiosis and nutrient acquisition. We transferred this trait to wheat, resulting in field-grown wheat with increased AM colonization and nutrient acquisition. Our findings reveal a new strategy to boost endosymbiosis in the field and reduce inorganic fertilizer use while sustaining plant growth. Nuclear calcium oscillations initiate plant–arbuscular mycorrhiza and nitrogen-fixing bacteria symbioses for nutrient acquisition, with a newly discovered autoactive CNGC15 mutant enhancing these partnerships, potentially improving crop nutrition and reducing inorganic fertilizer dependence.

|
Scooped by
The Sainsbury Lab
February 11, 3:30 PM
|
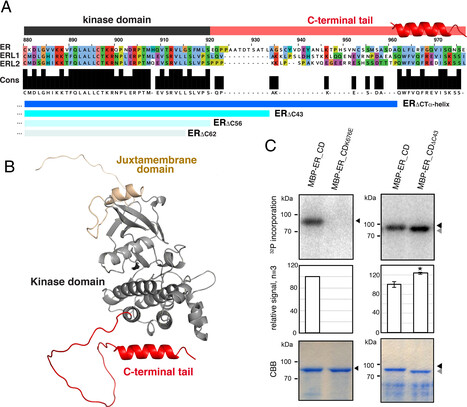
Cells perceive and process external signals through their cell-surface receptors, whose activity must be tightly maintained to prevent the spread of misinformation. How do plant cells prevent inappropriate receptor activity? We identify a structural module within the C-terminal tail of the receptor-kinase ERECTA (ER_CT) that inhibits the receptor pre- and post-signal activation. The ER_CT comprises a linker and an α-Helix. Before activation, ER_CT is autoinhibitory and associates with an inhibitory protein. Ligand perception triggers the transphosphorylation of ER_CT by the coreceptor, which then recruits a degradation machinery to turn over the activated receptor swiftly. Thus, we reveal an off–on–off toggle switch mechanism that finely adjusts the activity of the plant receptor, enabling precise control over cell signaling.

|
Scooped by
The Sainsbury Lab
February 11, 3:21 PM
|
Lichens are composite, symbiotic associations of fungi, algae, and bacteria that result in large, anatomically complex organisms adapted to many of the world’s most challenging environments. How such intricate, self-replicating lichen architectures develop from simple microbial components remains unknown because of their recalcitrance to experimental manipulation. Here, we report a metagenomic and metatranscriptomic analysis of the lichen Xanthoria parietina at different developmental stages. We identified 168 genomes of symbionts and lichen-associated microbes across the sampled thalli, including representatives of green algae, three different classes of fungi, and 14 bacterial phyla. By analyzing the occurrence of individual species across lichen thalli from diverse environments, we defined both substrate-specific and core microbial components of the lichen. Metatranscriptomic analysis of the principal fungal symbiont from three different developmental stages of a lichen, compared with axenically grown fungus, revealed differential gene expression profiles indicative of lichen-specific transporter functions, specific cell signaling, transcriptional regulation, and secondary metabolic capacity. Putative immunity-related proteins and lichen-specific structurally conserved secreted proteins resembling fungal pathogen effectors were also identified, consistent with a role for immunity modulation in lichen morphogenesis.

|
Scooped by
The Sainsbury Lab
December 12, 2024 11:47 AM
|
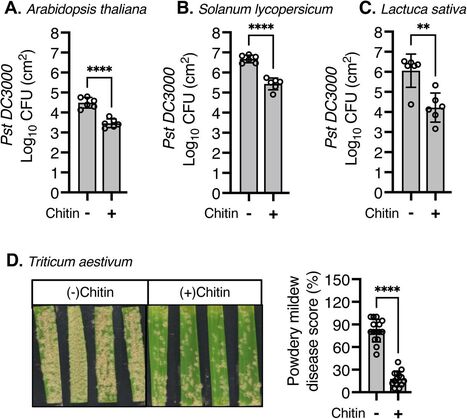
Chitin triggers localised and systemic plant immune responses, making it a promising treatment for sustainable disease resistance. However, the precise molecular mechanisms underlying chitin-induced systemic effects in plants remain unknown. In this study, we investigated the effects of soil amendment with crab chitin flakes (hereafter chitin) on pattern-triggered immunity (PTI) and systemic disease resistance in various plant species. We found that soil amendment with chitin potentiates PTI and disease resistance against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 in lettuce, tomato, and Arabidopsis as well as against the fungal pathogen Blumeria graminis causing powdery mildew in wheat. Using micrografting in Arabidopsis, we demonstrated that this systemic effect is dependent on active chitin perception in the roots. We also showed that induced systemic resistance (ISR) and pattern-recognition receptors (PRRs)/co-receptors, but not systemic acquired resistance (SAR), are involved in the systemic effects triggered by chitin soil amendment. This systemic effect correlated with the transcriptional up-regulation of key PTI components in distal leaves upon chitin soil amendment. Notably, chitin-triggered systemic immunity was independent of microbes present in soil or chitin flakes. Together, these findings contribute to a better understanding of chitin-triggered systemic immunity, from active chitin perception in roots to the potentiation of PTI in the leaves, ultimately priming plants to mount enhanced defense responses against pathogen attacks. Our study provides valuable insights into the molecular mechanisms of chitin soil amendment and resulting induced immunity, and highlights its potential use for sustainable crop protection strategies.

|
Scooped by
The Sainsbury Lab
December 12, 2024 11:26 AM
|
As much as 10% of plant immune receptors from the nucleotide-binding domain leucine-rich repeat (NLR) family carry integrated domains (IDs) that can directly bind pathogen effectors. However, it remains unclear whether direct binding to effectors is a universal feature of ID-containing NLRs given that only a few NLR-IDs have been functionally characterized. Here we show that the rice (Oryza sativa) sensor NLR-ID Pii2 confers resistance to strains of the rice blast fungus Magnaporthe oryzae that carry the effector AVR-Pii without directly binding this protein. First, we show that AVR-Pii binds the exocyst subunit OsExo70F2 in rice (Oryza sativa) to dissociate preformed complexes of OsExo70F2 with host RPM1 INTERACTING PROTEIN4 (RIN4) at the conserved NOI motif, facilitating a possible virulence function. Second, we show that in its resting state, Pii2 binds OsExo70F2 and OsExo70F3, essential components of Pii-mediated resistance, through its integrated NOI domain. Remarkably, AVR-Pii binding to OsExo70F2/F3 leads to dissociation of the Pii2–OsExo70F2 and Pii2–OsExo70F3 complexes, destabilization of Pii2, and activation of immunity. These findings support a novel conceptual model in which an NLR-ID monitors alterations of tethered host proteins targeted by pathogen effectors, providing insight into pathogen recognition mechanisms.

|
Scooped by
The Sainsbury Lab
December 12, 2024 10:04 AM
|
Rice blast disease, caused by the fungus Magnaporthe oryzae, is one of the most devastating diseases of rice. Therefore, understanding the mechanisms of blast fungus infection and resistance of rice against the disease is important for global food security. In this study, we show that the M. oryzae effector protein AVR-PikD binds rice sHMA proteins and stabilizes them, presumably to enhance pathogen infection. We show that loss-of-function mutants in one rice sHMA, OsHIPP20, reduced the level of susceptibility against a compatible isolate of M. oryzae, suggesting that M. oryzae requires host sHMA to facilitate invasion. Remarkably, OsHIPP20 knockout rice line showed no growth defect, suggesting editing sHMA genes may present a novel source of resistance against blast disease.

|
Scooped by
The Sainsbury Lab
November 18, 2024 5:21 PM
|
The emergence of fungal antimicrobial resistance—fAMR—is having a growing impact on human and animal health, and food security. This roadmap charts inter-related actions that will enhance our ability to mitigate the risk of fAMR. As humanity’s reliance on antifungal chemicals escalates, our understanding of their one-health consequences needs to scale accordingly if we are to protect our ability to manage the global spectrum of fungal disease sustainably.
|






 Your new post is loading...
Your new post is loading...