Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
July 7, 2021 7:32 AM
|
Highlights, news and events of I2BC in June 2021

|
Scooped by
I2BC Paris-Saclay
May 25, 2021 9:48 AM
|
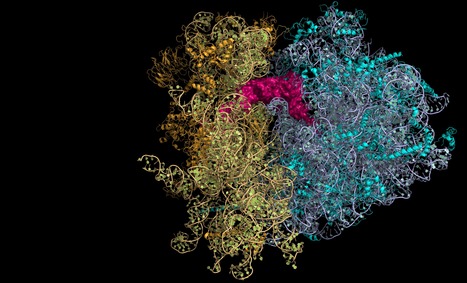
Translational accuracy of a tethered ribosome
What happens when the two subunits of the ribosome are attached together? Ribosomes universally consist of two independent subunits that assemble on the mRNA during translation initiation. Surprisingly, it was shown that it was possible by synthetic biology to covalently link the two subunits together. Although this is consistent with E. coli growth, the molecular consequences for translation fidelity were unknown. We therefore tested the functioning of this tethered ribosome, and were able to demonstrate that it is strongly impacted in its ability to recognise the SD sequence. In consequence, initiation and SD-dependent frame shifting are both impacted. We were also able to show that the translation termination step was less efficient. It is precisely at this step that the two subunits are physically dissociated. The fact that this is no longer possible in the tethered ribosome clearly disrupts the release efficiency of the ribosome.This work reveals the subtle perturbations linked to the attachment of the two ribosome subunits and should be taken into account in the biotechnological developments associated with the use of these tethered ribosomes https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab259/6266428#.YJLH79KSbnM.twitter Contact: olivier.namy@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 9, 2021 7:24 AM
|
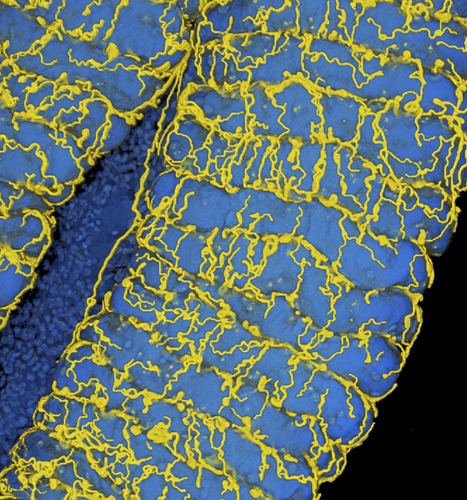
Oxygen transport to gut symbionts: a new route to fight insect pests?
Colonization of the insect gut with symbiotic bacteria triggers the development of a tracheal network that supplies oxygen to the symbionts. The insect respiratory system consists of tubular tracheae that transport oxygen to the organs. The tracheal network is dynamic and responds to developmental, environmental and nutritional cues. In a recent article published in the Proceedings of the National Academy of Sciences USA, the Plant Bacteria Interactions team of the Microbiology Department of I2BC, in collaboration with the team of Yoshitomo Kikuchi at the National Institute of Advanced Industrial Science and Technology – Hokkaido in Japan, shows that, in the insect pest Riptortus pedestris, the establishment of an essential symbiosis in the gut with the aerobic bacterial species Burkholderia insecticola triggers the development of an extensive tracheal network enveloping the gut. Genetically blocking the trachea formation prevents this gut symbiosis. The researchers further discovered that the reactive oxygen species-generating enzyme Duox is crucial for the formation and stabilization of tracheae by forming protein cross-links in the tracheal matrix. Reactive oxygen species generated by Duox can be scavenged with antioxidants such as N-acetylcysteine, and feeding insects with this compound prevents tracheal formation and symbiosis. Since many insects obligatorily depend on their symbioses, triggering their collapse by the specific inhibition of the respiratory network with antioxidants could be a new route to fight insect pests. More details here: https://doi.org/10.1073/pnas.2020922118 Contact: peter.mergaert@i2bc.paris-saclay.fr
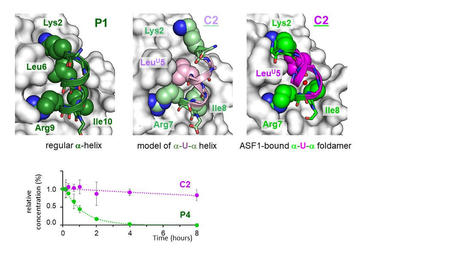
Dans une étude publiée dans Science Advances, des chercheurs de l’I2BC, en collaboration avec l'Institut Européen de Chimie et Biologie (Université Bordeaux, CNRS) ont développé une molécule chimérique de type peptide/foldamère, résistante à la protéolyse et inhibitrice d’ASF1, cible potentielle de traitement anticancéreux. La résolution de la structure de la chimère en interaction avec sa cible, première du genre, met en évidence une plasticité insoupçonnée du squelette à base d’urée qui épouse la surface d’ASF1 et maintient la même interface de liaison qu’un peptide inhibiteur non chimérique. Lire la suite de l'actualité CEA-Joliot ICI. Contact : francoise.ochsenbein@cea.fr
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
May 25, 2021 10:11 AM
|
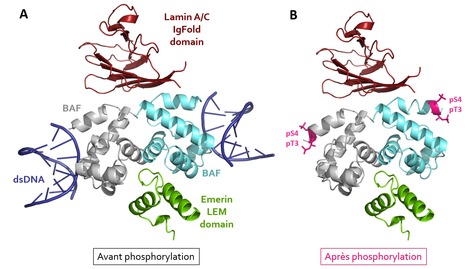
Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners
An NMR and X-ray cristallography study shows that mitotic phosphorylation of the BAF dimer considerably stiffens its structure, leads to its dissociation from DNA, but unexpectedly does not modify its binding to the nuclear envelope proteins emerin and lamin. In human cell nuclei, double-stranded DNA interacts with many proteins to form chromatin. Other proteins anchor chromatin to the nuclear membrane, which is important for genome stability. In 2018, our team elucidated the three-dimensional structure of a complex formed by the DNA-binding protein BAF, its inner nuclear membrane partner emerin, and a protein from the nucleoskeleton, lamin A/C. We showed that this complex is defective in patients with premature aging syndromes due to mutations in BAF or lamin A/C. It was reported that this complex must disassemble when cells divide (in mitosis), because then, the whole chromatin is reorganized. The present work aims at better understanding the mechanisms that regulate the assembly and disassembly of this complex during the cell cycle. Using biochemical, biophysical and structural biology experiments, we were able to reproduce in vitro the phosphorylation of the BAF protein by the mitotic kinase VRK1. We confirmed that BAF is doubly phosphorylated by VRK1 kinase, first on its serine 4, then on its threonine 3. These phosphorylation events significantly stiffen the N-terminal region of BAF, disrupt DNA binding, but unexpectedly, phosphorylated BAF remains bound to emerin and lamin A/C. This study paves the way for a broader exploration of the impact of mitotic phosphorylations on genome organization. Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners - PubMed (nih.gov) Contact: sophie.zinn@i2bc.paris-saclay.fr
Des chercheurs de l’I2BC en collaboration avec une équipe de l’institut Curie et de l’IRCM/CEA-Jacob, posent les bases moléculaires pour expliquer le double rôle du complexe Mlh1-Mlh3 dans la réparation des mésappariements d’ADN et, fait unique, dans l’une des étapes clefs du brassage génétique lors de la méiose. Leur étude est publiée dans PNAS. Lire la suite de l'Actu CEA-Jacob ICI. Contact : jb.charbonnier@cea.fr
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
May 25, 2021 11:24 AM
|
Bradyrhizobium atypical differentiation in Aeschynomene legumes
Multiomic analysis of a suboptimal symbiotic interaction where a soybean nodulator (Bradyrhizobium diazoefficiens) faces NCR peptides in Aeschynomene afraspera and undergoes atypical terminal bacteroid differentiation In the legume-rhizobium nitrogen-fixing symbiosis, some host plants produce antimicrobial peptides called NCR to control their bacterial partner and trigger its cellular differentiation. Consequently, intracellular bacteroids become polyploid, elongated and with a permeabilized membrane, collectively resulting in an altered bacterial viability. We call this phenomenon terminal bacteroid differentiation (TBD), which is associated to a higher return on investment to the host of the symbiotic process. In the study by Nicoud et al. published in mSystems, we analyzed the physiology of Bradyrhizobium diazoefficiens USDA110 in symbiosis with its natural host soybean, which does not trigger TBD, and in interaction with Aeschynomene afraspera, a host producing NCR peptides and inducing TBD. To do so, we combined whole-nodule metabolomics, with bacterial transcriptomics and proteomics. We found that the maladapted symbiosis between Bradyrhizobium and Aeschynomene is suboptimal (reduced benefit to the host) and that bacteria undergo an intense stress. Finally, we characterized bacteroid differentiation by a combination of flow cytometry and image based morphometry and found a disconnection of the canonical features of TBD, with bacteroids that fail to elongate and endoreduplicate, while their membranes become preamble and their viability reduced. https://doi.org/10.1128/mSystems.01237-20 Contact: benoit.alunni@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 9, 2021 7:39 AM
|
The Surviving Sepsis Campaign: Research Priorities for Coronavirus Disease 2019 in Critical Illness
The Surviving Sepsis Research Committee, a multiprofessional group of 17 international experts, identified important research priorities in the management, pathophysiology and host response of COVID 19 in critically ill patients. The Surviving Sepsis Research Committee provides 13 priorities for coronavirus disease 2019. Of these, the top six priorities were identified and include the following questions: 1) Should the approach to ventilator management differ from the standard approach in patients with acute hypoxic respiratory failure?, 2) Can the host response be modulated for therapeutic benefit?, 3) What specific cells are directly targeted by severe acute respiratory syndrome coronavirus 2, and how do these cells respond?, 4) Can early data be used to predict outcomes of COVID 19 and, by extension, to guide therapies?, 5) What is the role of prone positioning and noninvasive ventilation in nonventilated patients with coronavirus disease?, and 6) Which interventions are best to use for viral load modulation and when should they be given? Although knowledge of both biology and treatment has increased exponentially in the first year of the COVID 19 pandemic, significant knowledge gaps remain. The research priorities identified represent a roadmap for investigation in COVID 19. More details here: https://journals.lww.com/ccmjournal/Fulltext/2021/04000/The_Surviving_Sepsis_Campaign__Research_Priorities.5.aspx Contact: pierre.tissieres@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 10, 2021 8:14 AM
|
An innovative microfluidic system for mutation accumulation over many generations in budding yeast and genome-wide measurements of mutational profiles
Mutations in DNA have large-ranging consequences, from evolution to disease. Many mechanisms contribute to mutational processes such as dysfunctions in DNA repair pathways and exogenous or endogenous mutagen exposures. Model organisms and mutation accumulation (MA) experiments are indispensable to study mutagenesis. Classical mutation accumulation experiments with the plate cell culture method are labour intensive and time consuming. To fill the need for more efficient approaches to characterize mutational profiles, the groups of F. Malloggi (NIMBE/LIONS, IRAMIS, CEA) and J. Soutourina (Genome biology/I2BC, CEA/CNRS/Paris-Saclay) have developed and validated an innovative microfluidic-based system for automatizing MA culturing over many generations in budding yeast. This unique experimental tool, coupled with high-throughput sequencing, significantly streamlines and speeds up genome-wide measurements of mutational profiles, while also parallelizing and simplifying the cell culture. Indeed, this approach allows replacing manual colony plating – which requires 800 Petri dishes and human intervention every 2 days for more than 6 months – with a single microfluidic chip that contains up to 8 MA lines operating in parallel for 1–3 months with limited intervention. This makes it possible to simultaneously compare, in an unbiased manner, many yeast strains to precisely decipher genome-wide mutational processes. To validate our approach, we performed microfluidic MA experiments on two different genetic backgrounds, a wild-type strain and a base-excision DNA repair ung1 mutant characterized by a well-defined mutational profile. Crucially, we have demonstrated that the microfluidic approach also results in mutation accumulation comparable to the traditional experiment on plate. Our microfluidic approach will significantly improve MA experiments to study mutagenesis. Mutation accumulation analysis in model organisms such as budding yeast, where changes in DNA transactions and effects of environmental conditions can be precisely controlled, is important to understand mutational processes in eukaryotic cells. As most cellular functions are conserved from yeast to human cells, this knowledge can help to understand mutational processes at the origin of many human diseases including cancers and developmental disorders. Our approach thus paves the way to massively-parallel MA experiments with minimal human intervention that can be used to investigate mutational processes at the origin of human diseases and to identify mutagenic compounds relevant for medical and environmental research. More details here Contact : julie.soutourina@cea.fr

|
Scooped by
I2BC Paris-Saclay
May 25, 2021 10:05 AM
|
SAVE THE DATE - 2nd French congress on Integrative Structural Biology Paris-Saclay - November 29 to December 3, 2021
In love with Integrative Structural Biology? The next @bsi2021 meeting is the PLACE-TO-BE from nov 29-dec 3 2021. Co-organized by @AFC_cristal and SFBiophys societies in Paris-Saclay campus. Please save the date, follow bsi2021, check http://bsi-2021.cnrs.fr, and RT! The second French congress on Integrative Structural Biology (BSI), jointly organized by the Association Française de Cristallographie (AFC) and the Société Française de Biophysique (SFB), will take place on the Paris-Saclay campus from November 29 to December 3, 2021. The scientific program is available on the congress website. The congress can also be followed on twitter (@bsi2021). Structural biologists and biophysicists from I2BC (@I2BCParisSaclay), together with ENS-Paris-Saclay, Soleil, Ecole Polytechnic and ICSN are local organizers of the BSI202. https://bsi-2021.cnrs.fr/ Contact: julie.menetrey@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 19, 2021 3:38 PM
|
Hélène Bret, une thésarde du B3S, sélectionnée pour la demi-finale nationale de #MT180
Jeudi 11 mars 2021, l’ENS Paris-Saclay accueillait la finale Paris-Saclay du concours Ma Thèse en 180 seconde. Unique représentante de l'I2BC, Hélène BRET est arrivée deuxième. Bravo à elle et bonne chance pour la suite ! Quinze doctorants, issus d’une première sélection, ont présenté en 3 minutes leur sujet de thèse. Hélène Bret, étudiante dans l’équipe Assemblages macromoléculaires et intégrité des génomes (AMIG/I2BC) est arrivée deuxième de cette finale « régionale ». Elle a d’abord expliqué très simplement pourquoi il était important de savoir prédire les interfaces entre deux protéines et la tâche immense que cela représentait devant les millions de paires de protéines qui existent et le petit nombre d’interfaces déjà déterminées expérimentalement grâce aux méthodes de biologie structurale. Pendant sa thèse, Hélène construit un programme informatique qui doit prédire où est l’interface pour un couple de protéines. Ce programme est un « réseau de neurones artificiels » qui apprend tout seul à partir des exemples obtenus en laboratoire. Et Hélène lui apprend à se tromper de moins en moins. Hélène est ingénieur AgroParisTech spécialisée en science des données et apprentissage artificiel. Elle a obtenu en double diplôme un Master 2 ISI (Informatique : systèmes intelligents) de l'Université Paris-Dauphine. Elle est maintenant en deuxième année de thèse, co-encadrée par Raphaël Guérois et Jessica Andreani. Revoir sa prestation ici. En savoir plus à son sujet ici

|
Scooped by
I2BC Paris-Saclay
June 9, 2021 6:55 AM
|
Denise Zickler elected EMBO member 2021
We want to congratulate Dr. Denise Zickler for her election as an EMBO Member. Trained as a cytogeneticist, she has always been interested in the basic mechanisms that govern chromosome behavior during meiotic and mitotic divisions. Dr. Zickler has been a leader in the field since the 1980s and continues to push the community towards a better understanding of long-standing questions pertaining to homologous chromosome pairing and meiotic recombination. Her model system, the filamentous fungus Sordaria macrospora, became an unparalleled system to study meiosis. By clever genetic screens she identified all major players involved in the programmed DNA double-strand break formation that initiate meiotic recombination, ten to twenty years before their discovery in other organisms. 3D reconstruction of all seven chromosome pairs in each meiotic nucleus allowed to gain key insights into the pairing and recombination steps. This includes observation of the synaptonemal complex (SC), a proteinaceous structure between homologous chromosomes that stabilizes chromosome pairing and contributes to the regulation of crossover formation. By merging molecular, genetic and imaging analyses, Dr. Zickler has tackled the general problems of the connection between homologous pairing, SC initiation, crossover patterning with tremendous success (Cell, 2010; Genes Dev 2014; 2017; PNAS 2011; 2014). One of her last publications uncovered the existence of “bridges” between homologous chromosomes during the early stages of meiotic prophase. These bridges comprise structural components of the chromosome axes as well as recombination proteins. They allow to translocate the recombination proteins from the axes to the synaptonemal complex, mediating the interplay between this structure and the recombination process leading to crossover formation (PNAS 2019). She has been involved in many international collaborations, some that last to this day, bringing her intuition and her decisiveness to countless endeavors outside her own laboratory. Dr. Zickler always favored nurturing a small research group, wanting to stay nearly full time at the bench and believing in daily free exchanges of ideas. She has been an indispensable member of the meiosis community and, as such, has been an invited speaker at all major meiosis conferences for the last 20 years. She also spearheaded the creation of the EMBO Meiosis conference, that is still organized every other year and is a major event in the community. Her recognition as an EMBO member acknowledges her incomparable contribution over her fantastic career. Congratulations Denise! Additional information on EMBO announcement web page
|

|
Scooped by
I2BC Paris-Saclay
June 17, 2021 3:22 AM
|
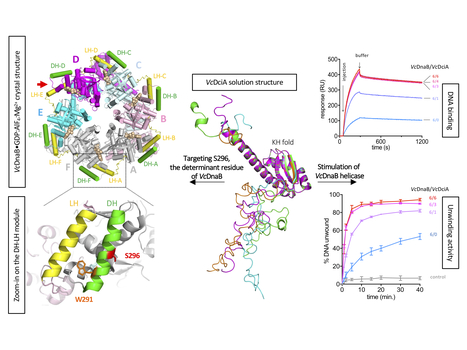
Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase
Replicative helicases are essential proteins that unwind DNA in front of replication forks. Their loading depends on accessory proteins and in bacteria, DnaC and DnaI are well characterized loaders. However, most bacteria do not express either of these two proteins. Instead, they are proposed to rely on DciA, an ancestral protein unrelated to DnaC/I. While the DciA structure from Vibrio cholerae shares no homology with DnaC, it reveals similarities with DnaA and DnaX, two proteins involved during replication initiation. As other bacterial replicative helicases, VcDnaB adopts a toroid-shaped homo-hexameric structure, but with a slightly open dynamic conformation in the free state. We show that VcDnaB can load itself on DNA in vitro and that VcDciA stimulates this function, resulting in an increased DNA unwinding. VcDciA interacts with VcDnaB with a 3/6 stoichiometry and we show that a determinant residue, which discriminates DciA- and DnaC/I-helicases, is critical in vivo. Our work is the first step toward the understanding of the ancestral mode of loading of bacterial replicative helicases on DNA. It sheds light on the strategy employed by phage helicase loaders to hijack bacterial replicative helicases and may explain the recurrent domestication of dnaC/I through evolution in bacteria. More details here Contacts: Sophie Quevillon-Cheruel@i2bc.paris-saclay.fr and Jean-Luc.FERAT@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 14, 2021 10:44 AM
|
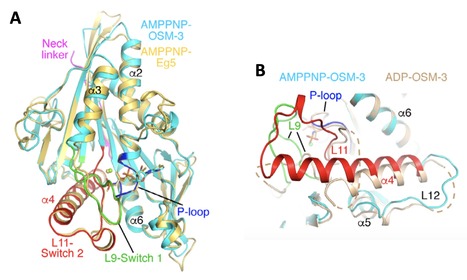
Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism
Motile kinesins are motor proteins that translocate along microtubules as they hydrolyze ATP. They share a conserved motor domain which harbors both ATPase and microtubule-binding activities. An ATP hydrolysis mechanism involving two water molecules has been proposed based on the structure of the kinesin-5 Eg5 bound to an ATP analog. Whether this mechanism is general in the kinesin superfamily remains uncertain. Here, we present structural snapshots of the motor domain of OSM-3 along its nucleotide cycle. OSM-3 belongs to the homodimeric kinesin-2 subfamily and is the Caenorhabditis elegans homologue of human KIF17. OSM-3 bound to ADP or devoid of a nucleotide shows features of ADP-kinesins with a docked neck linker. When bound to an ATP analog, OSM-3 adopts a conformation similar to those of several ATP-like kinesins, either isolated or bound to tubulin. Moreover, the OSM-3 nucleotide-binding site is virtually identical to that of ATP-like Eg5, demonstrating a shared ATPase mechanism. Therefore, our data extend to kinesin-2 the two-water ATP hydrolysis mechanism and further suggest that it is universal within the kinesin superfamily. More details here contact: julie.menetrey@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 21, 2021 5:35 AM
|
RNAi factors Dicer and Argonaute are essential for reproduction in the fungus Sordaria macrospora
While the ancestral role of the RNAi machinery in the eukaryotic ancestor(s) may have been geared toward targeting RNA transcript degradation and locus-specific histone modifications to promote gene silencing, findings in multiple organisms have now demonstrated the involvement of RNAi components in many other nuclear functions including centromere integrity, chromosome segregation and DNA repair. Previous findings implicated the RNAi machinery in the meiotic process, but left open the question of how these proteins impact the specific steps of prophase chromosome dynamics such as pairing, formation of the meiosis-specific structure called synaptonemal complex (SC), and homologous recombination. Here we exploited the power of Sordaria macrospora as a particularly attractive experimental system for examination of the roles of both Dicer and Argonaute proteins during its sexual cycle. We show that both Dicer proteins and both Argonautes present in Sordaria genome are dispensable for vegetative growth but required for the correct entry and progression into the sexual cycle, with a prominent role of one Dicer (Dcl1) and one Argonaute (Sms2). Moreover, we show that Dcl1, and in a lesser extent Dcl2 and the Argonaute Qde2, are involved in regulating chromosome axis length during meiosis, as well as crossover number and distribution along chromosomes. Taken together, these findings indicate clearly that the RNAi components play a central role in the meiotic process of Sordaria and extend our understanding of the different processes (e.g. axis length, crossover patterning) that are directly or indirectly dependent on the Dicer and/or the Argonaute proteins. More details here Contact: chloe.girard@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 9, 2021 5:44 AM
|
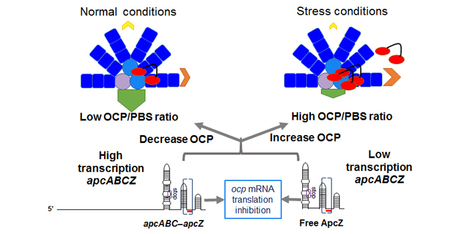
A small RNA linking light absorption and photoprotection
A posttranscriptional regulatory mechanism links the expression of the cyanobacterial Orange Carotenoid Protein related to photoprotection, directly and in an inverse fashion, to the synthesis of the phycobilisome, the cyanobacterial antenna via a 3’ end-derived sRNA. The modular photoactive Orange Carotenoid Protein (OCP), which has a crucial role in cyanobacterial photoprotection, has gained a great interest in the recent years due to its ability to act as a molecular photoswitch and to its suppressive 12 Å translocation of the carotenoid upon photoactivation. Upon blue light absorption, the inactive orange form (OCPO) converts to the active red state (OCPR) which is able to interact with the phycobilisome, the cyanobacterial antenna, and to decrease the energy arriving at the photochemical centers. Due to its function in thermal dissipation of excess energy, the expression of OCP must be tightly controlled to avoid a loss of energy under non-stressing conditions and to increase this dissipation under stressing ones. A research group of the I2BC in collaboration of a group of the University of Freiburg discovered the first molecular factor involved in the regulation of OCP expression. Moreover, they show for the first time that a sRNA appended to a long operon mRNA functions in the regulatory network of cyanobacteria. This factor is the first example in cyanobacteria of a sRNA derived from a polycistronic mRNA regulating at least one other mRNA in trans. The researchers demonstrated that ApcZ, a sRNA originating from the 3’end of the apcABC operon encoding the core phycobilisome proteins, is responsible for the repression of ocp translation under non-stress conditions. The transcription of the apcABC operon decreases under most stress conditions and as a consequence ApcZ (free and as part of the entire operon transcript) concentration decreases leading to a de-repression of ocp mRNA translation. Thus, light harvesting and photoprotection are connected directly and in an inverse fashion by a single regulatory sRNA. If, under stress conditions, less energy must arrive at the photochemical centers, the transcription of phycobilisome genes decreases and synthesis of OCP increases. Hence, the OCP concentration is controlled in a simple and elegant way. “Inverse Regulation of Light Harvesting and Photoprotection Is Mediated by a 3’ End-Derived sRNA in Cyanobacteria” Plant Cell (2021) 33, 358-380. Jiao Zhan, Claudia Steglich, Ingeborg Scholz, Wolfgang R. Hess and Diana Kirilovsky Contact: diana.kirlovsky@cea.fr

|
Scooped by
I2BC Paris-Saclay
June 14, 2021 10:47 AM
|
Observing a photoenzyme at work
Photoenzyme Fatty Acid Photodecarboxylase (FAP) recently discovered in microalgae is involved in light-driven generation of hydrocarbons from fatty acids, a process that bears great promise for the production of biofuels and the generation of high-value chemicals. The reaction mechanism has been elucidated in great detail using a combination of biochemical, time-resolved spectroscopic and crystallographic methods and theoretical approaches. More details here Contact: pavel.muller@i2bc.paris-saclay.fr
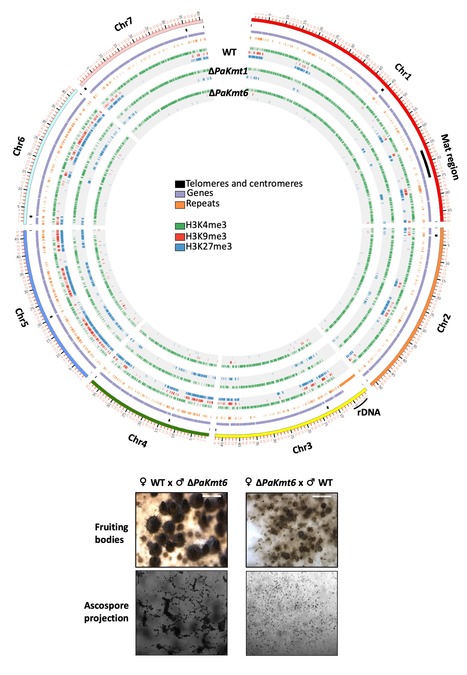
L'extinction sélective de l’expression des gènes est essentielle au développement. Chez les eucaryotes, il est généralement admis que l'hétérochromatine facultative enrichie en modifications H3K27me3 maintient la répression transcriptionnelle nécessaire aux processus de différenciation cellulaire. A l’inverse, l'hétérochromatine constitutive enrichie en modifications H3K9me3 est un frein à la différenciation cellulaire, mais protège les génomes de la mobilisation des éléments transposables qui les parasitent. Chez le champignon Podospora anserina, l’équipe de Fabienne Malagnac (I2BC, Gif-sur-Yvette) a établi les cartes génomiques des modifications H3K27me3 et H3K9me3 et constaté que ces marques s'excluent mutuellement dans les régions riches en gènes mais pas dans celles riches en éléments transposables. Les chercheurs ont ensuite inactivé les gènes codant les histone-méthyltransférases responsables de ces deux modifications. L'élimination de l'enzyme PaKmt6 EZH2-like entraîne non seulement une perte des modifications H3K27me3 mais aussi une réduction significative des modifications H3K9me3. De même, l'élimination de l'enzyme PaKmt1 de type SU(VAR)3-9 a entraîné une perte des modifications H3K9me3 et une diminution substantielle des modifications H3K27me3. Les chercheurs ont également établi que l'enzyme PaKmt6 est nécessaire au développement de P. anserina puisque son absence provoque de graves défauts dans la plupart des séquences de son cycle de vie, tandis que la perte de l'enzyme PaKmt1 n'entraîne que des défauts marginaux. Ces résultats, publiés dans Epigenetics & Chromatin, montrent une fonction conservée du complexe répresseur Polycomb (PRC2) dans le développement fongique, mais suggèrent une fluidité évolutive intrigante dans la machinerie de dépôt des marques d’histones, qui remet en question les définitions canoniques de l'hétérochromatine constitutive et facultative. Contact : pierre.grognet@universite-paris-saclay.fr ou fabienne.malagnac@universite-paris-saclay.fr
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
May 25, 2021 9:59 AM
|
The key lipopolysaccharide modifications for antibiotic resistance in Escherichia coli
Escherichia coli acquires resistance to polymyxin B upon the activation of two-component systems, but this is at the expense of innate bile acid resistance. The lack of lipopolysaccharide phosphorylation accounts for bile susceptibility and impairs intestinal colonization in mice. The surface properties of bacterial cells play a key role in resistance to various antimicrobial agents. Gram-negative bacteria expose lipopolysaccharides (LPS) on their surface, which can undergo different structural modifications. However, some of these modifications may be beneficial in some circumstances, but detrimental in others. In this study, published in Frontiers in Microbiology, the team of Thierry Touzé, in collaboration with a team from Institut Pasteur, showed that Escherichia coli cells resist to cationic antimicrobial peptide polymyxin B, one last resort antibiotic, under conditions that simultaneously activate two-component regulatory systems PmrA/B and PhoP/Q. Among a set of modifications, they identified those responsible for this resistance, which occurs on the lipid A moiety (known as the endotoxin) from LPS. The acquisition of polymyxin B resistance came at the expense of loss of innate resistance to deoxycholate, a major component of bile. They provide evidences that bile susceptibility arises from the inhibition of LpxT-dependent modification, which consists in lipid A phosphorylation. Bile acids are abundant in the small intestine, where they modulate the commensal flora and the researchers further showed that the inactivation of lpxT impaired gut colonization in mice. These results highlight the importance of lipid A decorations and their tight regulation in the lifestyle of E. coli and probably other Enterobacteriaceae that exhibit the same modifications https://www.frontiersin.org/articles/10.3389/fmicb.2021.676596/full Contact: thierry.touze@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 9, 2021 7:28 AM
|
SEPSIS, le vrai visage de la COVID-19 : Mieux comprendre et mieux soigner
Dans le cadre de la Journée mondiale de lutte contre le sepsis, le 13 septembre 2021, la Fédération Hospitalo Universitaire (FHU) SEPSIS organise le symposium "SEPSIS, le vrai visage de la COVID-19 : Mieux comprendre et mieux soigner". Ce symposium se déroulera au Ministère des Solidarités et de la Santé (Amphithéâtre Laroque- 14, avenue Duquesne 75007 Paris). La matinée comprendra des interventions destinées plus particulièrement aux scientifiques, alors que l’après-midi sera consacrée à l’information du grand public par des conférences de vulgarisation. Pour participer, il est nécessaire de vous inscrire en ligne. Le programme et le bulletin d'inscription sont disponibles à l'adresse ci-dessous: https://www.fhu-sepsis.uvsq.fr/symposium-sepsis-le-vrai-visage-de-la-covid-19-mieux-comprendre-pour-mieux-soigner Contact : pierre.tissieres@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 19, 2021 8:36 AM
|
Microfabrication room opening in I2BC
We are glad to announce the opening of the microfabrication room in Gif-sur-Yvette (room 00-120, Bâtiment 23). This equipment was funded by “Région Ile de France” under the DIM ELICIT (Domaine d’Intérêt Majeur “Empowering LIfe sCiences with Innovative Technologies”) Microfluidics is the science of fluids manipulation at the microscale using channels with dimensions of tens to hundreds of micrometers. It is also a technology called Lab-on-Chip used in Physics, Chemistry and Biology. The properties of flows at the microscale allow to work faster and cheaper with smaller sample volumes. The polymer used to produce microfluidics chip is transparent and biocompatible and provides access to time-lapse live imaging of biological systems on a microscope. It has many applications like cells sorting, encapsulation of drugs, DNA sequencing, micro-organisms swimming, mixing, etc.
The microfabrication plateform is a clean room fully equiped to make microfluidic chips from the mold design and its characterization, to the polymer microchip fabrication. It is managed by Jessica MARION et Vassanti AUDEMAR who handle the equipment :
- Clewin 5 : software to design the chip
- Laminator (Peak) or spin coater (Laurell) and UV exposer (Kloé) : to produce molds
- Profilometer (Filmetrics) : to characterize the mold
- Plasma cleaner (Harrick Plasma) : to seal the microfluidic chip
We will be happy to welcome you.
Contacts : Sébastien THOMINE (sebastien.thomine@i2bc.paris-saclay.fr), Jessica MARION ( jessica.marion@i2bc.paris-saclay.fr ), Vassanti AUDEMAR ( vassanti.audemar@i2bc.paris-saclay.fr )

|
Scooped by
I2BC Paris-Saclay
May 28, 2021 3:39 AM
|
Introduction à la cristallographie biologique
Dans le monde du vivant, les mécanismes mis en jeu pour respirer, digérer, grandir ou bouger un bras, impliquent des molécules constituées de milliers d’atomes, ce sont les macromolécules. Ces macromolécules circulent, interagissent et se contorsionnent en permanence. Visualiser ces macromolécules à l’échelle atomique constitue une aide incomparable pour comprendre comment elles fonctionnent, comment elles dysfonctionnent, et pour concevoir des médicaments, inhibiteurs ou activateurs. Quand on utilise la structure tridimensionnelle d’une macromolécule, il est essentiel de garder un œil critique, et pour cela, il faut comprendre comment cette structure a été obtenue. La cristallographie aux rayons X est une méthode historique qui utilise des rayons X et des cristaux pour déterminer la structure tridimensionnelle des molécules à l’échelle atomique. Elle est également la méthode la plus répandue. Le livre « Introduction à la cristallographie biologique » s’adresse d’abord aux biologistes, mais aussi à toute personne intéressée par la biologie structurale. Les auteurs vous proposent une initiation aux différentes étapes de la cristallographie biologique qui mène de la cristallisation à la structure tridimensionnelle d’une macromolécule. Ce livre fait suite au MOOC « Voyage au cœur du vivant avec des rayons X : la cristallographie ». Il peut en être le compagnon ou être utilisé seul. Grace aux vidéos accessibles par les codes QR de chaque fin de chapitre, des lieux d’ordinaire fermés au public sont ouverts pour vous offrir un voyage unique au cœur du vivant. Marie-Hélène le Du est biophysicienne au CEA à l’Institut de Biologie Intégrative de la Cellule (I2BC/CNRS) de l’Université Paris-Saclay. Pierre Legrand est scientifique de ligne sur la ligne Proxima-1 au Synchrotron SOLEIL. Serena Sirigu est scientifique de ligne sur la ligne Proxima-2A au Synchrotron SOLEIL. Sylvain Ravy est directeur de recherche au CNRS, et directeur du Laboratoire de Physique des Solides (LPS/CNRS) de l’Université Paris-Saclay. Introduction à la cristallographie biologique - De Marie-Hélène Le Du, Pierre Legrand, Serena Sirigu et Sylvain Ravy (EDP Sciences) Contact : marie-helene.ledu@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
June 25, 2021 11:47 AM
|
Portrait of a young researcher from the I2BC: Joana Santos, genome biology department
Joana Santos starts her own team at the I2BC. Joana Santos joined the Genome Biology Department of the I2BC (UMR 9198-CNRS, CEA, Universite Paris-Saclay), as lab head of the team” Transcription Regulation of the Malaria Parasite” in January 2020. In October 2020, she became a CNRS CRCN. Joana Santos graduated with a Bachelor in Genetics and Microbiology from the University of Lisbon in 2005. Fascinated by parasites, she then moved to the US, where she did a one-year internship in the laboratory of Dr Photini Sinnis, at NYU, NY. There, she studied how the rodent malaria parasite, Plasmodium berghei, invades the host liver cells. Pursuing her interest in mechanisms of host cell invasion by parasites, she moved, in 2006, to Geneva, Switzerland, to undergo a PhD under the supervision of Prof Dominique Soldati, as part of the Marie-Curie Early Training PhD Program MalPar. Her work focused on an important family of parasite ligands that play roles post-host-cell attachment, in a signaling pathway, that may activate a transcription program. This got her interested in transcription regulation in parasites so in 2011, she moved to the lab of Prof Manuel Llinás, at Princeton University, NJ, USA, to study how the human malaria parasite P. falciparum, activates transcription of the genes allowing invasion of the host red blood cell. In 2017, she did a second postdoc, now in Toulouse, France, in the lab of Dr Amine Khamlichi, at the IPBS (UMR 5089 – CNRS, Universite Paul Sabatier) to deepen her knowledge of transcription regulation mechanisms, now in B cells. In 2019, Joana Santos was awarded an ATIP-Avenir grant, which allowed her to start her own team at the I2BC. Her team focuses on transcriptional and post-transcriptional regulation mechanisms of the human malaria parasite. The work developed in her team will allow a better understanding of the malaria parasite, which continues to cause the death of over 400 thousand people in the world, yearly, and the discovery and characterization of new drug targets.
|

 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...