Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 6:04 AM
|
Polo-like kinase 1 (Plk1) regulates replication origin activation and interacts with Rif1
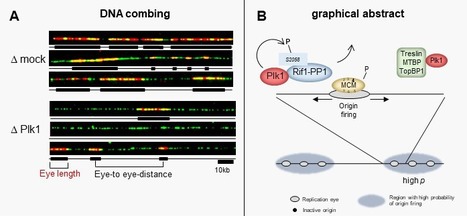
Using different approaches, the group of K. Marheineke in collaboration with A. Goldar (J. Soutourina group) and the proteomics platforms at I2CB and at Necker hospital found a new mechanism of how the kinase Plk1 promotes DNA replication via inhibiting the replication repressor Rif1. The activation of eukaryotic DNA replication origins needs to be strictly controlled at multiple steps in order to faithfully duplicate the genome and to maintain its stability. How the checkpoint recovery and adaptation protein Polo-like kinase 1 (Plk1) regulates the firing of replication origins during non-challenged S phase remained an open question. Using DNA fiber analysis, the group of Kathrin Marheineke in collaboration with Arach Goldar (J. Soutourina lab) show that immunodepletion of Plk1 in the Xenopus in vitro system decreases replication fork density and initiation frequency. Numerical analyses suggest that Plk1 reduces the overall probability and synchrony of origin firing. We used quantitative chromatin proteomics and co-immunoprecipitations in collaboration with SiCAPS I2BC proteomics platform to demonstrate that Plk1 interacts with firing factors MTBP/Treslin/TopBP1 as well as with Rif1, a known regulator of replication timing. Phosphopeptide analysis by LC/MS/MS show that the C-terminal domain of Rif1, which is necessary for its repressive action on origins through protein phosphatase 1 (PP1), can be phosphorylated in vitro by Plk1 on S2058 in its PP1 binding site. The phosphomimetic S2058D mutant interrupts the Rif1-PP1 interaction and modulates DNA replication. Collectively, our study provides molecular insights into how Plk1 regulates the spatio-temporal replication program and suggests that Plk1 controls origin activation at the level of large chromatin domains in vertebrates. More details here Contact persons: Kathrin Marheineke & Arach Goldar Follow the teams of Kathrin Marheineke and of Arach Goldar

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 5:18 AM
|
Revisiting the multiscale structuring of bacterial chromosomes
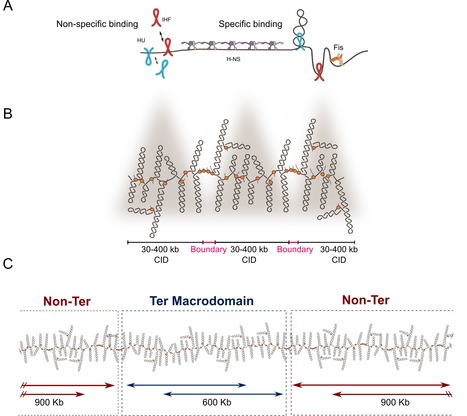
Do you know how bacteria organize their chromosomes allowing key biological processes for the cell? Here we review old and new factors that are essential for the structuring of the bacterial chromosome. During the last decades, works have shown that bacterial chromosomes are not just molecules of DNA inside a bag composed of lipids, peptidoglycan and proteins. Recent advances have not only confirm the multiscale organization of bacterial chromosome, but also have added new layers on chromosome organization. Briefly, DNA supercoiling is organized into stochastic 10-kb domains included in larger 40–300-kb chromosomal interaction domains (CIDs) delimited by long and highly expressed gene clusters. The bacterial chromatin composed of multiple nucleoid associated proteins (NAPs) bound to DNA not only influences gene expression but also modulates, together with bacterial SMC complexes, the folding and disposition of the chromosome in the cell. Although the main structural layers and their key organizers start to emerge, the complete picture of chromosome organization in bacteria remains elusive. In this article, researchers of the Genome Biology Department of the I2BC review not only the classical well-known factors involved in chromosome organization but also novel components that have recently been shown to dynamically shape the 3D structuring of the bacterial genome. More details here Contact person: Frédéric Boccard Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 4:54 AM
|
A disordered cryptic repeat in BRCA2 binds to a newly discovered meiotic protein
The interaction between BRCA2 and its interactor HSF2BP, both required for meiotic recombination, is defined structurally, revealing that a repeat in BRCA2 binds two HSF2BP units, increasing the affinity up to the nanomolar range. Unexpectedly, this repeat is not essential for mouse meiosis. BRCA2 and its interactors are required for meiotic homologous recombination (HR) and fertility. Loss of HSF2BP, a BRCA2 interactor, disrupts HR during spermatogenesis. We test the model postulating that HSF2BP localizes BRCA2 to meiotic HR sites, by solving the crystal structure of the BRCA2 fragment in complex with dimeric armadillo domain (ARM) of HSF2BP and disrupting this interaction in a mouse model. This reveals a repeated 23 amino acid motif in BRCA2, each binding the same conserved surface of one ARM domain. In the complex, two BRCA2 fragments hold together two ARM dimers, through a large interface responsible for the nanomolar affinity — the strongest interaction involving BRCA2 measured so far. Deleting exon 12, encoding the first repeat, from mBrca2 disrupts BRCA2 binding to HSF2BP, but does not phenocopy HSF2BP loss. Thus, results herein suggest that the high-affinity oligomerization-inducing BRCA2-HSF2BP interaction is not required for RAD51 and DMC1 recombinase localization in meiotic HR. More details here Contact person : Sophie Zinn Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 4:09 AM
|
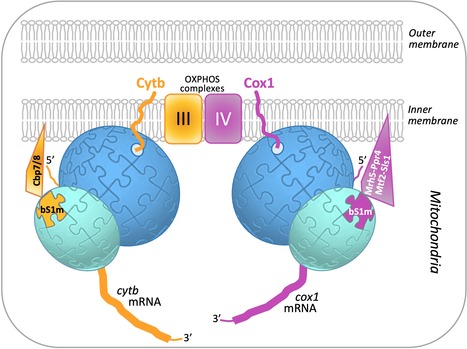
Translational specificity mediated by mitoribosomal isoforms
A combination of genetic screens and mass spectrometry analyses has revealed that an interplay of translational activators and a heterogeneous mitoribosome population balances the production of respiratory complex subunits in fission yeast mitochondria. Mitochondria contain their own DNA and translation machinery. Mitochondrial mRNAs encode key subunits of the oxidative phosphorylation (OXPHOS) complexes, that produce energy for the whole cell. Thus, mitochondrial translation defects lead to severe diseases in humans. The fission yeast Schizosaccharomyces pombe is a valuable model to study mitochondrial gene expression, since it closely resembles humans in its mitochondrial DNA structure and physiology. By combining bioinformatics, genetic and biochemical approaches including mass spectrometry on the I2BC SICaPS platform, the I2BC BIOMIT group discovered two interacting factors of S. pombe, Cbp7 and Cbp8, controlling the production of Cytb, a catalytic subunit of OXPHOS complex III. Two classes of Cbp7/Cbp8 partners were identified and shown to modulate the synthesis of Cytb or Cox1, key subunits of the OXPHOS complexes III and IV respectively. First, two isoforms of bS1m, a protein of the small mitoribosomal subunit, that appear mutually exclusive and confer translational specificity. Second, a complex of four proteins dedicated to Cox1 synthesis, which includes an RNA helicase, Mrh5, that interacts with the mitochondrial ribosome. These data suggest that S. pombe contains, in addition to complexes of translational activators, a heterogeneous population of mitochondrial ribosomes that could specifically modulate translation depending on the mRNA translated, in order to optimally balance the production of different respiratory complex subunits. Together, these results, published in Nucleic Acids Research, support the view that ribosomes are not merely translation machines, but can also behave as regulatory elements. More details here Contact person : Nathalie Bonnefoy Follow the team
Avec les Domaines de Recherche et d'Innovation Majeurs (DRIM), la Région poursuit son action en faveur de projets de recherche structurants portés par des équipes pluridisciplinaires autour de thématiques scientifiques d'intérêt majeur. Pourquoi : Pour renforcer l’attractivité et l’excellence scientifique et technologique de la recherche en Île-de-France dans des disciplines et thématiques d’intérêt majeur pour l’Île-de-France. Pour quoi ? Pour labelliser et financer des réseaux de recherche structurants et fédérateurs d’équipes de recherche et d’entreprises, porteurs de programmes de recherche et d’innovation sur la période 2022-2026 (estimation : 2,5 M€ maximum par an par réseau).
Pour qui ? Pour des équipes de recherche de toutes disciplines souhaitant porter des projets pluridisciplinaires et fédérer des réseaux de recherche franciliens autour d’enjeux scientifiques et d’innovation.
Comment candidater ? Avant le 14 octobre 2021 à 17h00, depuis la plateforme régionale : https://mesdemarches.iledefrance.fr Plus d'informations ICI.
Via Life Sciences UPSaclay
Ce prix récompense des chercheurs ayant mené, avec une PME ou une entreprise de taille intermédiaire, un partenariat de recherche ayant eu un réel impact économique (augmentation du chiffre d'affaires, création d'emplois).
Date limite de candidature : 21 octobre 2021
Via Life Sciences UPSaclay
La Région Ile-de-France est bénéficiaire d'un projet européen COFUND : Paris Region Fellowship, dont l'objectif est de financer des post-doctorats de 24 mois. Les postes peuvent profiter à des projets rentrant dans le cadre des 11 DIMs, mais pas uniquement - il est vivement conseillée scruter les conditions d'éligibilité des jeunes chercheurs qui seront recrutés.
Voir https://parisregionfp.sciencescall.org/ Date limite le 21 Octobre, 2021 à 17:00
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
July 27, 2021 5:35 AM
|
Plant endosymbionts defend themselves against the hostile host environment
Rhizobium endosymbionts of legume plants use multiple mechanisms for resistance against antimicrobial peptides produced by the host plant cells. The nitrogen fixing symbiosis of legumes with rhizobium bacteria has a predominant ecological role in the nitrogen cycle and has the potential to provide the nitrogen required for plant growth in agriculture. The host plants allow the nitrogen-fixing rhizobia to colonize the cells of specific symbiotic organs, the nodules, in very large numbers in order to produce sufficient reduced nitrogen for the plant needs. Some legumes, including Medicago spp., produce massively antimicrobial peptides to keep this large bacterial population in check. These peptides, known as NCRs, have the potential to kill the rhizobia but in the nodule cells, they rather inhibit the division of the endosymbionts and trigger them into a morphologically differentiated state, resulting in a high nitrogen fixing activity. In this study published in mBio, the Plant-Bacteria Interactions team of I2BC shows that the bacterial resistance to the antimicrobial activity of the NCR peptides in the Medicago symbiont Sinorhizobium meliloti is multifactorial and requires peptide transporters, the lipopolysaccharide outer membrane and the stress response regulator RpoH1. More details here Contact: peter.mergaert@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:59 AM
|
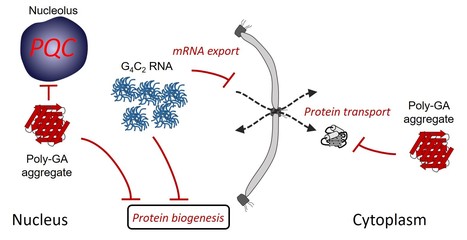
Multiple pathways of toxicity induced by C9orf72 dipeptide repeat aggregates and G4C2 RNA in a cellular model
The RNA and protein products from C9orf72 mutations causing amyotrophic lateral sclerosis and frontotemporal dementia induce various and additive proteostasis impairments. The most frequent genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia is a G4C2 repeat expansion in the C9orf72 gene. This expansion gives rise to translation of aggregating dipeptide repeat (DPR) proteins, including poly-GA as the most abundant species. However, gain of toxic function effects have been attributed to either the DPRs or the pathological G4C2 RNA. Here, we analyzed in a cellular model the relative toxicity of DPRs and RNA. Cytoplasmic poly-GA aggregates, generated in the absence of G4C2 RNA, interfered with nucleocytoplasmic protein transport, but had little effect on cell viability. In contrast, nuclear poly-GA was more toxic, impairing nucleolar protein quality control and protein biosynthesis. Production of the G4C2 RNA strongly reduced viability independent of DPR translation and caused pronounced inhibition of nuclear mRNA export and protein biogenesis. Thus, while the toxic effects of G4C2 RNA predominate in the cellular model used, DPRs exert additive effects that may contribute to pathology. More details here Contact: frederic.frottin@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:44 AM
|
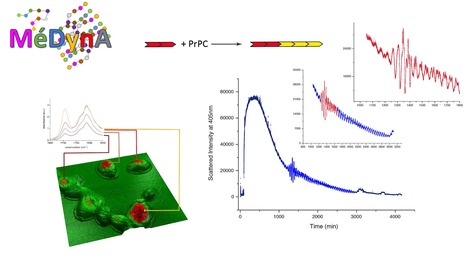
Second MéDynA plenary meeting (Assembly mechanisms and dynamics of self-organised protein-based complexes)
The IMAPP group is prominently involved in the MéDynA second plenary meeting, 6-9 September, 2021 at Ile d'Oléron (organising committee: Sonia Fieulaine, Stéphane Bressanelli, Maïté Paternostre, Edith Godard, Yves Boulard). MéDynA is a 'GdR' (coordination of labs all across France around a specific research topic) whose objectives are to: * Bring together French labs studying by diverse means the dynamical pathways leading to self-organised protein assemblies or protein-based assemblies (incl. peptidomimetics)
* Forge a common language so as to benefit from the multiple expertise stemming from B13https://medyna.cnrs.fr/les-actualites/deuxieme-pleniere/B14 biophysics, chemistry, physics and mathematics to understand and master mechanisms of protein assembly
* Foster emergence of interdisciplinary and novel research projects
* Set up a nationwide interdisciplinary network useful to all researchers, whether at an early career stage (such as graduate students and postdocs) or well established More information here Contact: gdr.medyna@ibpc.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:23 AM
|
Plantes - Dissiper l’énergie lumineuse en excès sous forme de chaleur pour se protéger : lumière sur les mécanismes moléculaires
De CEA-Joliot.fr : Dans une étude publiée dans JBC, une équipe de l'I2BC, en collaboration avec l’Institut botanique de Beijing (Chine), décrit pour la première fois un état intermédiaire de l’antenne collectrice de lumière des plantes supérieures qui permet de mieux comprendre les changements qui s’opèrent au niveau moléculaire lors de l’activation d’un mécanisme de photoprotection de la plante. La photosynthèse commence par l’absorption de l’énergie lumineuse par les pigments chlorophylles des antennes collectrices de la lumière (LHC- Light Harvesting Complex, complexes composés de protéines et de pigments chlorophylles et caroténoïdes). Cette absorption crée une énergie d’excitation (passage d’un état électronique fondamental à un état excité de la chlorophylle collectrice), énergie qui est transférée de proche en proche, d’une chlorophylle à un autre, jusqu’au centre réactionnel de la photosynthèse où elle est convertie en énergie de potentiel chimique (par séparation de charge). Ce processus de conversion de l’énergie est extrêmement efficace. Tellement efficace, qu’il peut provoquer une surexcitation du système potentiellement délétère. La plante met en place des mécanismes pour s’en prémunir : de l’échelle macroscopique, par le mouvement de ses feuilles, à l’échelle moléculaire, par un mécanisme qui permet la dissipation de l’énergie sous forme de chaleur. Ce dernier mécanisme, multifactoriel, est appelé extinction non photochimique de la fluorescence de la chlorophylle (non photochemical quenching).
Que ce soit in vivo ou in vitro, l’équipe Bioénergétique Membranaire et stress (département I2BC), dirigée par Bruno Robert, a montré que ce quenching est lié à un réarrangement des protéines et pigments qui composent le LHC qui crée des « pièges à « énergie ». Les pigments chlorophylles excités transfèrent leur énergie à des pigments caroténoïdes qui la dissipent immédiatement sous forme de chaleur. Dans le cas du LHCII, la principale antenne collectrice des plantes supérieures, des expériences de spectroscopie in vitro menées sur le complexe agrégé (en l’absence de détergent utilisé classiquement pour le solubiliser) établissent que ce transfert se fait entre la chlorophylle a et une lutéine (un caroténoïde). L'ampleur de l'extinction semble corrélée aux changements de conformation (torsion) affectant la lutéine et un autre caroténoïde, la néoxanthine.
Pour aller plus loin dans la description et la compréhension de ces modifications, l’équipe de Bruno Robert, a étudié la structure du LHCII dans différents environnements qui influencent ses propriétés électroniques. De manière remarquable, l’équipe a réussi à isoler pour la première fois un état de LHCII, obtenu en utilisant le détergent n-dodécyl-α-D-maltoside, et à caractériser les propriétés spectroscopiques de ses pigments. Leur étude, publiée dans JBC, montre que, dans cet état, sont présents tous les changements associés à l’extinction non photochimique (changements dans les interactions protéine-chlorophylle, torsion de la néoxanthine) à l’exception de la torsion de lutéine, alors qu’aucun quenching n’est associé à cet état. Cet état de LHCII serait en quelque sorte un état intermédiaire qui permettrait le passage d’un état « allumé », capable d’absorber et de convertir l’énergie lumineuse en énergie chimique, à un état « éteint » par l’extinction non photochimique. La torsion de la néoxanthine serait un indicateur de changements de conformation à grande échelle du LHCII qui précéderaient des changements à plus petite échelle directement à l’origine du quenching et revélés par la torsion de la lutéine. Cet intermédiaire LHCII non éteint, décrit ici pour la première fois, permet de mieux comprendre le mécanisme moléculaire de l'extinction. More details here Contact : Bruno.ROBERT@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:52 AM
|
La science ouverte, un défi pour la recherche et pour le CEA
De joliot.cea.fr : Depuis quelques années, les orientations de la Commission européenne en faveur de la science ouverte et le Plan national pour la science ouverte en France incitent les chercheurs à rejoindre ce mouvement. Le CEA soutient depuis 2008 le libre accès aux publications scientifiques avec la création de son portail HAL-CEA et renforce aujourd’hui son engagement pour la science ouverte en publiant sa « Charte pour une science ouverte ». Explications avec Elsa Cortijo, directrice de la recherche fondamentale au CEA et présidente du comité de pilotage de l’information scientifique et technique du CEA. Lire la suite de l'Actualité CEA-Joliot ici
Pour déposer des publications sur le portail HAL, de nombreux tutoriels sont à la disposition des chercheurs ici:
https://doc.archives-ouvertes.fr/tutoriels-video/

|
Scooped by
I2BC Paris-Saclay
July 5, 2021 8:56 AM
|
Structural Bioinformatics Mini-Symposium on 8th July @JOBIM2021
The field of structural bioinformatics aims at developing algorithms to tackle the current challenges in 3D structure prediction of biological macromolecules and their functional annotation by exploiting data from genomics and biophysics in combination with systems biology and design, to be applied in biotechnology and therapeutics. This mini-symposium proposes to focus on integrative structural modeling and to address the new challenges, in a redefined landscape owing to the recent developments in experimental biophysics on one hand (the resolution revolution in cryo-EM) and in deep-learning on the other hand (as demonstrated by the extraordinary performances of AlphaFold2 unveiled in December 2020). These rapid changes are a sign of acceleration in structural bioinformatics developments such as structural modeling using cryo-EM or other experimental data, progress in machine learning for structural prediction and protein design. This mini-symposium will occur on 8th July 2021 and feature talks by Riccardo Pellarin, Ezgi Karaca, Sergei Grudinin and Slavica Jonic. It is co-organized by Jessica Andreani, Gwenaëlle André-Leroux, Benjamin Bardiaux, Stéphanie Baud, Isaure Chauvot de Beauchêne, Elodie Laine and Juliette Martin. JOBIM registration is required to attend the mini-symposium. More details here Contact: Jessica.ANDREANI@i2bc.paris-saclay.fr
|

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 5:37 AM
|
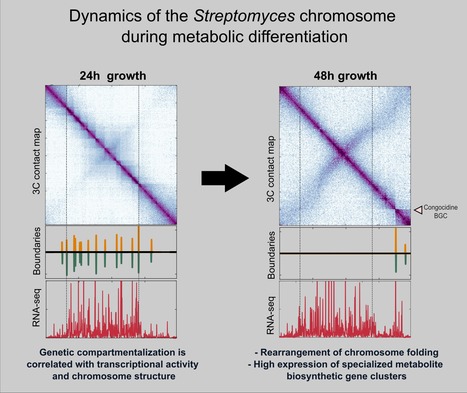
The folding dynamics of Streptomyces' chromosome during metabolic differentiation is revealed
A joint collaborative effort of the Microbiology, Genome Biology Departments and the Sequencing Facility of the I2BC unveiled the dynamics of the linear chromosome of Streptomyces ambofaciens, during metabolic differentiation. Streptomyces are soil bacteria mostly known for their complex life cycle (uni- to multi-cellular transition, sporulation, metabolic differentiation) and their prolific specialized metabolism (e.g. antibiotics and pigments), widely exploited in medicine, agriculture and the food industry. Their genome possesses remarkable characteristics including a large size (6-12 Mb) and a high GC content (circa 72%) and, even more unusual in bacteria, a linear configuration. Interestingly, Streptomyces chromosomes present a remarkable genetic compartmentalization, with a distinguishable central region harboring core genes and two terminal regions enriched in specialized metabolite biosynthetic gene clusters. The molecular mechanisms governing the structure and function of these compartmentalized chromosomes remain mostly unknown. In this work, teams of the Genome Biology, the Microbiology Department and the Sequencing Facility of the I2BC in collaboration with the DynAMIC (Lorraine University -INRAe) and the TIMC-IMAG ( Grenoble Alpes University - CNRS) Institutes, show that chromosome structure in Streptomyces ambofaciens correlates with genetic compartmentalization during exponential phase. Conserved, large and highly transcribed genes form boundaries that segment the central part of the chromosome into domains, whereas the terminal ends tend to be transcriptionally quiescent compartments with different structural features. The onset of metabolic differentiation is accompanied by a rearrangement of the chromosome architecture, from a rather ‘open’ to a ‘closed’ conformation, in which highly expressed specialized metabolite biosynthetic genes form new domain boundaries. Altogether, these results indicate that the linear chromosome of S. ambofaciens is partitioned into structurally distinct entities, suggesting a link between chromosome folding, gene expression and genome evolution. More details here Contact persons: Virginia Lioy & Stephanie Bury-Mone Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 5:12 AM
|
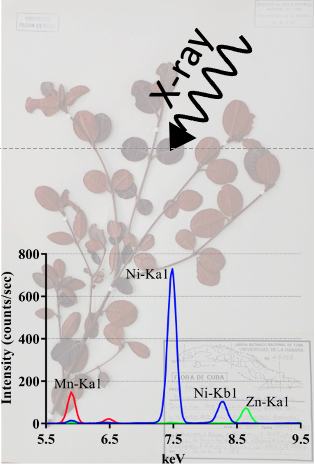
The X-ray fluorescence screening of multiple elements in herbarium specimens from the Neotropical region reveals new records of metal accumulation in plants
X-ray analysis of herbaria specimens from the Neotropical region provides an overview of plant metal accumulation and reveals new Mn, Zn and Ni hyperaccumulators.
Plants have developed a diversity of strategies to take up and store essential metals in order to colonize various types of soils including mineralized soils. Yet, our knowledge of the capacity of plant species to accumulate metals is still fragmentary across the plant kingdom. In this study, we have used the X-ray fluorescence technology to analyze metal concentration in a wide diversity of species of the Neotropical flora that was not extensively investigated so far. In total, we screened more than 11 000 specimens representing about 5000 species from herbaria in Paris and Cuba. Our study provides a large overview of the accumulation of metals such as manganese, zinc, and nickel in the Neotropical flora. We report 30 new nickel hyperaccumulating species from Cuba, including the first records in the families Connaraceae, Melastomataceae, Polygonaceae, Santalaceae, and Urticaceae. We also identified the first species from this region of the world that can be considered as manganese hyperaccumulators in the genera Lomatia (Proteaceae), Calycogonium (Melastomataceae), Ilex (Aquifoliaceae), Morella (Myricaceae), and Pimenta (Myrtaceae). Finally, we report the first zinc hyperaccumulator, Rinorea multivenosa (Violaceae), from the Amazonas region. The identification of species able to accumulate high amounts of metals will become instrumental to support the development of phytotechnologies in order to limit the impact of soil metal pollution in this region of the world. More details here Contact person: Sylvain Merlot Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 4:36 AM
|
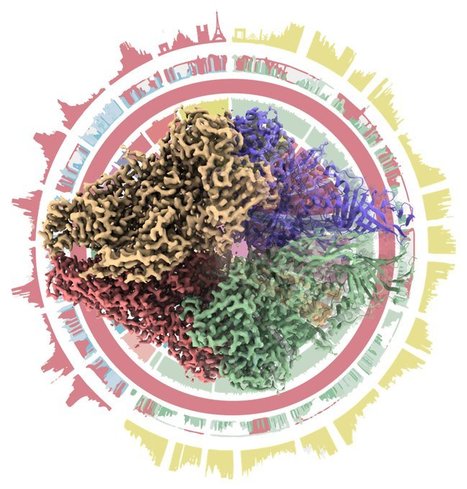
CryoEM of NHEJ supercomplex
The study reports the CryoEM structures of the core proteins involved in the human NHEJ pathway that repair most DNA double-strand breaks generated in human cells. Double strand breaks (DSB) are the most toxic DNA lesions in the cell. The Non-homologous end-joining pathway is the main DNA repair pathway in mammals that recognizes, process and ligates DSB. It is initiated by the recognition of DSB by the heterodimer Ku70/80 and the kinase DNA-PKcs that can form a synapse between the two DNA ends of the DSB. Ku70/80 then recruits several enzymatic activities to process and ligate the DNA ends. A team from B3S I2BC collaborates with TL Blundell laboratory (University of Cambridge) to determine cryoEM structure at 4.3Angstrom of the synapse formed by Ku70/80, DNA-PKcs, the ligation complex Ligase4/XRCC4/XLF. This super-complex show a central role of XLF that bridges Ku70/80 and Ligase4/XRCC4 on both side of the synapse. Mutations of the dimer interfaces negatively affect DNA repair. More details here Contact person : Jean-Baptiste Charbonnier Follow the team
La Fondation Nouveaux Espoirs lance son premier appel à candidatures pour financer des projets de recherche sur la maladie de Lyme et les maladies vectorielles à tiques. Date limite de candidature le 22 octobre 2021.
Via Life Sciences UPSaclay
Cet appel vise à initier et renforcer des coopérations bilatérales entre la France et la Finlande en recherche, innovation et enseignement supérieur. Date limite de candidature : 31 octobre 2021.
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
July 26, 2021 8:28 AM
|
Zeiss on your campus truck !
For more than a year, the unprecedented context in which we live has prevented any meetings, a brand new format of physical event where Zeiss come to meet you, as close as possible to your place of work ! Tradeshows and conferences are ideal opportunities to discover new products and witness innovations firsthand. A real-time experience in an individual hands-on session will be offer to you to test new functionalities and give you the opportunity to talk to an expert about your needs. With Zeiss on your campus Truck,you can have the opportunity to experience microscopy workflows at first hand. Near your place of practice and taking into account the applicable hygiene measures, you can individually test the ZEISS instruments and solutions. More details here Contact : Sandrine.LECART@i2bc.paris-saclay.fr
Cet appel d'offres, lancé en partenariat par l'Inserm et le CNRS, vise à permettre à de jeunes chercheurs de mettre en place et d'animer une équipe au sein d'une structure de recherche Inserm ou CNRS. ATIP-Avenir Call 2022 (EN)
AO Atip-Avenir 2022 (FR) Opening of the registration online : October 18th 2021
Deadline for the online submission : November 18th 2021
Preselection : Mid-April 2022
Interviews : Mid-June 2022 Documents to download You may submit all the documents completed along with the online application. You will find practical information to access and fill the application in the Guide for applicant 2022. Contacts
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:48 AM
|
Temporal compartmentalization of viral infection in bacterial cells
Membrane-less compartments are generated by viral infection of a bacterial cell. Virus lytic infection imposes a major biosynthetic effort to the host cell and takes over significant cellular space. Viruses of prokaryotes must meet the challenge to restructure the cytoplasm open space of a small-sized cell. A study published in the Proceedings of the National Academy of Sciences USA reports the discovery that bacteriophage SPP1 infection leads to biogenesis of two types of membraneless compartments in the cytoplasm of the bacterium Bacillus subtilis. One is a single viral DNA compartment and others are warehouses for storage of viral particles. These compartments are temporal and spatially independent.
The DNA compartment sequesters machines operating viral DNA transactions. Multiple hybrid DNA replication centers, containing both phage replication proteins and hijacked bacterial replisomes, operate at different sub-locations within the compartment for parallelized synthesis of viral DNA. DNA is subsequently packaged in virion precursors without DNA (procapsids) at the compartment edges. Viral DNA-filled capsids then segregate from the DNA compartment, bind phage tails, and the resulting virions build warehouse compartments.
This spatial partition of the B subtilis cell responds to the requirements for exponential replication of SPP1 genomes and for the assembly of hundreds of viral particles. Its similarities to remodelling of the cell nucleus by herpesviruses led to the hypothesis that ancestral strategies used by viruses to invade the cell space were conserved to infect hosts of different Domains of Life.
Part of this work is from the PhD thesis of Audrey Labarde at Université Paris-Saclay. More details here Contact: paulo.tavares@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:37 AM
|
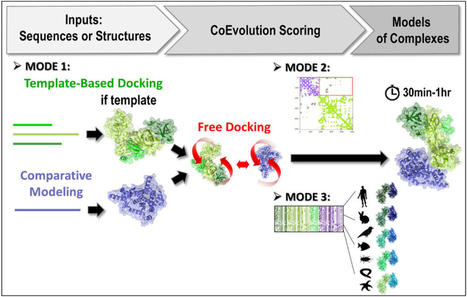
3rd version of the InterEvDock server: better exploiting protein sequence evolution data to improving the prediction of interface structures
De CEA-Joliot.fr : Researchers at I2BC, in collaboration with the RPBS platform, have developed the third version of their InterEvDock server for structural modelling of protein-protein interactions. The server integrates new algorithms for exploiting sequence evolution information. This greatly improves its performance for the generation of correct assembly models. Predicting the structure of proteins and their interaction modes is a real challenge for structural biologists. In addition to the gap between the number of proteins whose sequence is known and the number of available structures, proteomics has recently revealed the previously hidden part of the iceberg: hundreds of thousands of physical interactions between proteins. Knowing the surfaces involved in these interactions is essential, not only for understanding the mechanisms that govern how cells and organisms function, but also for designing new therapeutic or enzymatic molecules for pharmaceutical and biotechnological purposes.
Jessica Andreani and Raphaël Guérois (Team “Molecular Assemblies and Genome Integrity”/LBSR/I2BC) have been working for several years on the modelling of protein-protein interactions. In particular, they contribute to the improvement of prediction methods by integrating an evolutionary dimension into molecular docking tools. Indeed, protein interfaces tend to be more conserved than other regions on the protein surface. Moreover, signs of co-evolution can be detected at interfaces, where potentially disruptive mutations are compensated for by mutations in contacting positions on the protein partner. The team has thus developed and released the InterEvDock server, in collaboration with the Ressource Parisienne en Bioinformatique Structurale (University of Paris). As in previous versions, the InterEvDock3 server offers a systematic search for possible interfaces between two partners (known as free docking) and generates numerous conformations that are then ranked, in particular by taking into account information on the evolution of protein sequences. This unique modelling server processes user requests in a variety of formats (structural or sequence data, on one or both partners).
The software is now in its third version (InterEvDock3). This version integrates three new prediction modes that are described in two articles published in NAR1 and Bioinformatics2. The first mode, which is not based on free docking, allows homology modelling of large complexes with potentially low sequence identity. It uses sequences (no structures) as input and runs a template-based modelling protocol by searching exhaustively for close and distant homologs to generate assembly models.
The second mode predicts the structure of complexes from contact maps resulting from methods combining covariation analysis and deep learning. This mode uses 3D structures of monomers or homomultimers (such as a helicase hexamer) to perform a free docking approach while trying to satisfy the contacts predicted in the contact map. It is able to handle some ambiguous information, especially if one of the two partners is a homomultimer (with its residues thus present several times in the structure) and a contact in the predicted map can thus materialise in different ways.
Finally, the third mode uses 3D structures of monomers or multimeric complexes (possibly modelled from sequences by mode 1) and implements a new strategy for evaluating interfaces with coevolutionary information. Ten to forty representative pairs of homologous sequences (i.e. ten to forty evolutionarily conserved interactions between homologs) are selected, modelled at the atomic scale and scored. This new algorithm, tested on a database of 752 complexes (see Bioinformatics2), increases the number of correctly predicted complexes by 30%.
It usually takes between 20 and 60 minutes for the server to propose an interaction model. Developed with funding from two national health and biology infrastructures (FRISBI and IFB), the server is accessible from the RPBS platform:
https://bioserv.rpbs.univ-paris-diderot.fr/services/InterEvDock3/ Encart sur Proteo3Dnet
Another server to analyze interaction data obtained by proteomics
The team also collaborates with the RPBS on the Proteo3Dnet3 server designed to analyze interactions identified by proteomic techniques by integrating structural information (in particular 3D structures of known complexes). Developed with the funding of three national infrastructures in health and biology (FRISBI, ProFI and IFB), it is accessible from the RPBS: https://bioserv.rpbs.univ-paris-diderot.fr/services/Proteo3Dnet/ More details: https://doi.org/10.1093/nar/gkab358 https://doi.org/10.1093/bioinformatics/btab254 https://doi.org/10.1093/nar/gkab332 Contact: Jessica.ANDREANI@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:15 AM
|
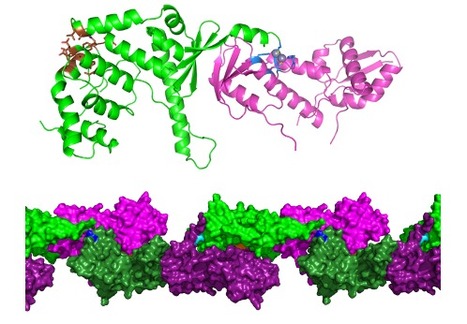
Molecular basis of the “scissors” mechanism involved in meiotic recombination and DNA repair
De CEA-Joliot.fr : Researchers from I2BC in collaboration with a team from Institut Curie and IRCM/CEA-Jacob, lay the molecular basis to explain the dual role of the Mlh1-Mlh3 complex in DNA mismatch repair and, quite uniquely, in one of the key steps of genetic mixing during meiosis. Their study has been published in PNAS. During meiosis, recombination of genetic material between homologous chromosomes occurs, which contributes to genetic mixing. Recombination can only take place through fine mechanisms of breakage and subsequent repair of DNA molecules. These mechanisms are highly conserved in eukaryotes, from yeast to humans. In particular, cross-shaped structures (Holliday junctions) are formed between homologous chromosomes to make crossovers during recombination. These recombination mechanisms are essential to ensure correct segregation of chromosomes in gametes. Failure of these steps results in the generation of gametes with an abnormal number of chromosomes (trisomy, Turner syndrome, infertility).
The Mlh1-Mlh3 complex is a repair factor for DNA mismatches that are generated as a result of errors in replicative polymerases. Mlh1-Mlh3 has endonuclease activity, i.e. it can cut a DNA molecule between two successive nucleotides and not at its ends. Unlike other similar mismatch repair factors, Mlh1-Mlh3 also exerts its endonuclease activity during meiosis, at the Holliday junctions; it is essential for the exchange of chromosome portions. The Mlh1-Pms1 complex, which is the main factor for repairing DNA mismatches, is not involved in meiosis. However, the two complexes have many structural similarities. For example, they are formed by the interaction between the C-terminal domains of Mlh1 and of its partner Mlh3 or Pms1. What are the structural bases for the differences in specificity between Mlh1-Mlh3 and Mlh1-Pms1? Researchers from I2BC B3S department, Team Nuclear Enveloppe, Telomeres and DNA repair) and the Institut Curie, with the help of CEA-Jacob (IRCM department) and a Swiss team, have solved the three-dimensional structure of a complex formed by the interaction domains of Mlh1 and Mlh3 (from purified recombinant proteins) of the yeast S. cerevisiae by radiocrystallography, and have functionally characterized it. They compared it with the already known equivalent complex formed between Mlh1 and Pms1.
Their study, published in PNAS, reveals differences between the two complexes, particularly with regard to the size of the heterodimerisation interface. The regulatory domains of Pms1 and Mlh3 are oriented differently in the complex with Mlh1. In addition, the shape of the cavity surrounding the endonuclease site varies, which could lead to differences in specificity towards their DNA substrates. The last ten residues of Mlh1 are known to be essential for mismatch repair but not for interaction with Pms1 or Mlh3. Experiments with mutant S. cerevisiae strains - into which deletions of the last residues of Mlh1 have been introduced - indicate that only the last 3 residues are essential for the meiotic activity of Mlh1. Other (DNA binding) experiments allow the authors to conclude that Mlh1-Mlh3, but not Mlh1-Pms1, binds preferentially to Holliday junctions. Finally, in the crystal, the Mlh1-Mlh3 dimers associate with each other to form a filamentary structure. This conformation supports the hypothesis proposed in previous studies that Mlh1-Mlh3 is oligomerised along the DNA. Mutations in the corresponding interaction surfaces strongly decrease the formation of chromosome crossovers. The authors of the study propose that Mlh1-Mlh3 oligomerization starts at a Holliday junction before extending away along the DNA.
This first comparison at the structural level of the Mlh1-Mlh3 and Mlh1-Pms1 complexes strongly suggests an evolutionary specialization of each. To go further, it will be necessary to obtain cryo-electron microscopy structures of the two whole proteins, in interaction with their DNA substrate. More details here Contact : jb.charbonnier@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 19, 2021 8:07 AM
|
Accueil des doctorants de l'I2BC
L'I2BC a enfin pu accueillir en présentiel ses nouveaux doctorants… enfin ceux qui ont débuté leur doctorat en octobre 2020! Le 29 juin dernier, l'I2BC a enfin pu réunir les doctorants qui ont débuté leur doctorat en automne dernier. Cette promotion est arrivée dans l'unité à la veille de confinements successifs, ce qui a hélas retardé cet accueil initialement prévu en novembre 2020. Les doctorants ont reçu un sweatshirt blanc aux logo de l'I2BC et de l'université Paris-Saclay d’où leur nom de « promotion Blanche ». La direction leur a présenté l'ensemble de l'unité. En effet, souvent confinés dans leurs équipes respectives pendant leur doctorat, ils découvrent probablement trop tardivement les autres départements et équipes de recherche ainsi que les immenses offres technologiques proposées par les plateformes de l'unité. Cette présentation générale de l’unité s’est poursuivie par une présentation de l’association « YourI2BC » ainsi que d’un pot d’accueil. Contact : maite.paternostre@i2bc.paris-saclay.fr ou frederic.boccard@i2bc.paris-saclay.fr
|



 Your new post is loading...
Your new post is loading...