Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
December 10, 2025 4:16 AM
|
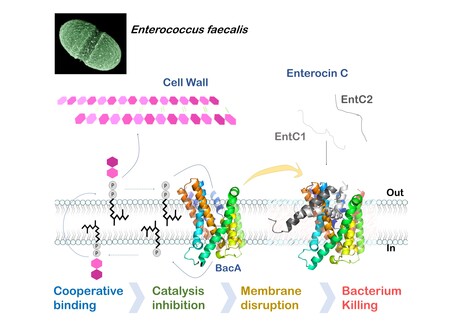
Mechanistic insights into Enterocin C bacteriocin targeting Enterococcus faecalis
Hijacking mechanism of membrane undecaprenyl phosphate recycling protein BacA in Enterococcus faecalis by a two-peptide bacteriocin revealed Antibiotic-resistance is a critical global health concern stressing the urgent need for new therapeutic strategies beyond conventional drugs. In this context, peptide-based bacteriocins constitute potential medical use antibacterial alternatives. In the present study, we have highlighted the molecular mechanism of a two-peptide bacteriocin, enterocin C, with potent activity against Enterococcus faecalis, a major opportunistic pathogen notorious for multidrug resistance. In collaboration with a group from INRAE MICALIS, the team has uncovered how enterocin C specifically exploits the membrane-embedded protein BacA as a cell surface receptor. BacA, widely conserved across bacteria, plays a central role in bacterial cell-wall biogenesis through its dual phosphatase and flippase activities, which are essential for recycling the lipid carrier undecaprenyl phosphate.
Using a combination of biochemical, biophysical, and microbiological approaches supported by AlphaFold structural modelling, we dissected the cooperative action of the two enterocin C peptides. Acting at nanomolar concentrations, peptide EntC1 inserts deeply into BacA’s outward-open catalytic pocket, blocking its enzymatic function and facilitating the subsequent binding of peptide EntC2. This dual docking event anchors the bacteriocin deep within the membrane’s hydrophobic core, ultimately triggering membrane disruption and bacterial cell death.
The findings reveal the molecular determinants of this precision targeting and represent the first detailed mechanistic description of a two-peptide bacteriocin’s mode of action. This work identifies BacA as a valuable target for bacteriocin-mediated killing and open avenues for the rational design of peptide-based antimicrobials for tailor-made antimicrobials to help combat antibiotic-resistant infections. More information : Thierry Touzé :thierry.touze@i2bc.paris-saclay.fr https://pubmed.ncbi.nlm.nih.gov/41232670/

|
Scooped by
I2BC Paris-Saclay
December 8, 2025 4:54 AM
|
Selective elimination of donor bacteria enables global profiling of plasmid gene expression during conjugation
A new ED-TA method which enabled genomics investigation of plasmid establishment during conjugative transfer was developed by I2BC researchers. They showed robust induction of a subset of plasmid genes at the early stages of conjugation through single-stranded DNA promoters. Bacterial conjugation is a principal mode of horizontal gene transfer which has important life science implications including bacterial genome evolution, dissemination of genetic traits and bioengineering applications. Notably, the spread of multidrug resistance via conjugative plasmids is one of the biggest concerns in global public health. Although conjugation has been studied since 1940s and the overall procedure is widely known and well documented, molecular details of reactions that establishes a transferred plasmid in the new host cell remain elusive. In addition, there are specific regulatory mechanisms for temporal expression of plasmid genes, that are also crucial for successful conjugation as they allow timely expression and function of plasmid-encoded arsenal against host defense mechanisms. Genomics-based studies of plasmid establishment were previously hampered by the nature of conjugation which takes place within a mixture of cell populations. Essentially, they require the separation of subpopulations before the DNA/RNA extraction to avoid contamination of indistinguishable plasmid DNA/RNA from the donor. Researchers of the Genome Biology Department of the I2BC, described the development of a new method, called ED-TA, which exploits a donor mutant hypersensitive to hypoosmotic shock. ED-TA allows unprecedently quick and efficient « Elimination of Donor population for Transconjugant Analysis». Using a clinically relevant model multidrug resistant conjugative plasmid, pESBL, they elucidated its transcription profile in successful and abortive conjugation. Researchers also showed single-stranded DNA promoters allow robust induction of a subset of genes at the early stages of conjugation. As the ED-TA method is straightforward and broadly applicable, further research taking advantage of the method will shed light on important molecular mechanisms of plasmid establishment after conjugation. More information: doi.org/10.1093/nar/gkaf1299 https://www.insb.cnrs.fr/fr/cnrsinfo/une-nouvelle-methode-pour-suivre-de-pres-la-transmission-des-genes-chez-les-bacteries Contact: Yoshiharu Yamaichi yoshiharu.yamaichi@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 1, 2025 4:49 AM
|
The Biophysics facility welcomes Viola Caroline D'mello
The Biophysics facility is pleased to welcome Viola Caroline D’mello, who joined in March 2025 as a CEA Ingénieure Chercheuse. After obtaining her Bachelor’s degree in Chemistry from Mumbai University and her Master’s in Analytical Chemistry from Mangalore University, she pursued her doctoral studies at the Tata Institute of Fundamental Research (TIFR) in Mumbai. Her thesis focused on the gas-phase study of hydrogen bonds in nitrogen-containing aromatic molecules using nanosecond UV and IR spectroscopy — hydrogen bonds similar to those found in DNA and RNA. In 2019, she joined LIDYL research group at Institute IRAMIS-CEA Saclay as a postdoctoral researcher, where she studied ion pairs and peptide folding in the gas phase. After a short period of working in industry in Bangalore, India, she returned to academia as a postdoctoral scholar in the Department of Physics at the University of Gothenburg, where she contributed to the commissioning of a high-resolution mass spectrometer coupled to a supersonic jet source. Since joining the Biophysics facility, she has taken charge of the electronic, Raman, and infrared spectroscopy facilities, overseeing user access while ensuring that experiments are designed and conducted under optimal conditions. She routinely supports and advises users on a wide range of techniques, including UV–Vis absorption, ultrafast transient absorption (from femtoseconds to milliseconds), FTIR, and Raman spectroscopy. Her broad expertise in photophysical methods, together with her reliability and open, collaborative attitude, makes her a central pillar of the Biophysics facility. More information : https://www.i2bc.paris-saclay.fr/biophysics/
Contact : biophysics@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
November 13, 2025 7:31 AM
|
The PIM facility welcomes Anaëlle and a new instrument: the Prometheus PANTA
A new piece of equipment was recently acquired by the Macromolecular Interactions Measurement facility (PIM) at the I2BC: the Prometheus PANTA, developed by Nanotemper Technologies. The Prometheus PANTA combines several detection methods to simultaneously measure thermal stability and aggregation tendency. Its technology is mainly based on nanoDSF (Differential Scanning Fluorimetry), which detects changes in protein conformation as a function of heating temperature, without the need for fluorescent labeling. The intrinsic fluorescence of aromatic amino acids (tryptophan and tyrosine) serves as a probe to track thermal transitions and determine denaturation points (Tm), which are direct indicators of protein stability. In addition, the system incorporates Dynamic Light Scattering (DLS) to measure hydrodynamic radius and detect oligomer formation, as well as a Back Reflection measurement that provides information on solution turbidity and protein aggregation. These combined measurements provide a global view of protein behavior under different experimental conditions, with precise temperature control (from 15 to 110°C) and excellent reproducibility. Each analysis requires only a few microliters of sample (10µL per capillary). The Prometheus PANTA is therefore a versatile tool for studying biomolecules. It allows you to: -
Evaluate the thermal stability of proteins -
Study interactions between proteins and a ligand -
Optimize buffer formulation, purification, and storage conditions -
Monitor the formation of aggregates or condensates under varying temperature or composition conditions. Its sensitivity, speed of analysis, and ability to measure multiple samples, up to 48 capillaries in parallel, make it a tool suitable for both fundamental research and biotechnology or structural biology projects. The arrival of Prometheus PANTA is accompanied by that of Anaëlle KALUBI KABAMBI, an assistant engineer recently recruited to the PIM facility. She will be responsible for implementing and optimizing analysis protocols, as well as assisting users in sample preparation and data interpretation. Her expertise in biochemistry will strengthen the facility’s capabilities, helping to provide researchers with comprehensive support for characterizing the interactions and structural properties of biomolecules Complementing the other instruments available at PIM (DSC, ITC, BLI, FIDA, and MST), the Prometheus PANTA enhances the facility’s ability to offer a complete range of protein characterization techniques. More information: https://www.i2bc.paris-saclay.fr/structural-biology/pim/ Contacts: anaelle.kalubi-kabambi@i2bc.paris-saclay.fr ; stephanie.marsin@i2bc.paris-saclay.fr ; magali.aumont-nicaise@i2bc.paris-saclay.fr ; magali.noiray@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 15, 2025 9:07 AM
|
Moss BRCA2 lacking the canonical DNA-binding domain promotes homologous recombination and binds to DNA
Despite having low homology with BRCA2 proteins from other organisms and no folded domain, the newly discovered BRCA2 protein from Physcomitrium patens (the green yeast) promotes homologous recombination by binding to recombinases and DNA. BRCA2 is crucial for mediating homology-directed DNA repair (HDR) through its binding to single-stranded DNA (ssDNA) and the recombinases RAD51 and DMC1. Most BRCA2 orthologs have a canonical DNA-binding domain (DBD) with the exception of Drosophila melanogaster. It remains unclear whether such a noncanonical BRCA2 variant without DBD possesses a DNA-binding activity. Here, we identify a new noncanonical BRCA2 in the model plant Physcomitrium patens (PpBRCA2). We establish that PpBRCA2 is essential for genome integrity maintenance, somatic DNA double-strand break (DSB) repair, HDR-mediated gene targeting, and RAD51 foci recruitment at DNA break sites. PpBRCA2 is also critical for DSB repair during meiosis. Interestingly, PpBRCA2 interacts strongly with RAD51 but weakly with DMC1, suggesting a distinct meiotic function compared to other BRCA2 homologs. Despite lacking the canonical DBD, PpBRCA2 binds ssDNA through its disordered N-terminal region and efficiently promotes HDR. Our work highlights that the ssDNA binding capacity of BRCA2 homologs is conserved regardless of the presence of a canonical DBD and provides a deeper understanding of BRCA2’s functional diversity across species. More information: https://pmc.ncbi.nlm.nih.gov/articles/PMC12412785/ Contact: Sophie Zinn sophie.zinn@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 5, 2025 3:50 AM
|
Restriction of Ku translocation protects telomere ends
How Ku inward translocation can be restricted to protect telomere ends from NHEJ without altering other important functions. Safeguarding chromosome ends against fusions via nonhomologous end joining (NHEJ) is essential for genome integrity. Paradoxically, the conserved NHEJ core factor Ku binds telomere ends. How it is prevented from promoting NHEJ remains unclear, as does the mechanism that allows Ku to coexist with telomere-protective DNA binding proteins, Rap1 in Saccharomyces cerevisiae. Here, we find that Rap1 directly inhibits Ku’s NHEJ function at telomeres. A single Rap1 molecule near a double-stand break suppresses NHEJ without displacing Ku in cells. Furthermore, Rap1 and Ku form a complex on short DNA duplexes in vitro. Cryo-EM shows Rap1 blocks Ku’s inward translocation on DNA – an essential step for NHEJ at DSBs. Nanopore sequencing of telomere fusions confirms this mechanism protects native telomere ends. These find- ings uncover a telomere protection mechanism where Rap1 restricts Ku’s inward translocation. This switches Ku from a repair-promoting to a protective role preventing NHEJ at telomeres. More information : https://doi.org/10.1038/s41467-025-61864-1 Contact : Philippe Cuniasse philippe.cuniasse@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
August 20, 2025 3:34 AM
|
Confocal microscopy workshop 2025

|
Scooped by
I2BC Paris-Saclay
June 30, 2025 5:14 AM
|
Training session on TEM in cell biology - 14-17 October 2025

|
Scooped by
I2BC Paris-Saclay
May 7, 2025 4:30 AM
|
New training session on FIB-SEM applications to biology at ambient temperature

|
Scooped by
I2BC Paris-Saclay
April 15, 2025 8:44 AM
|
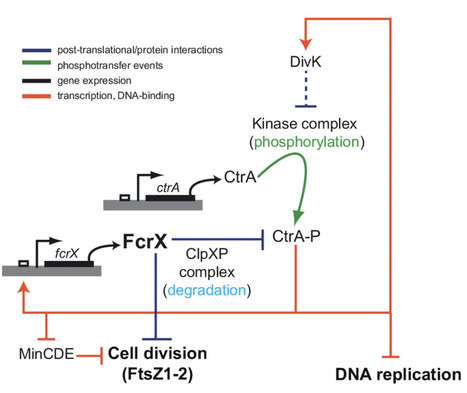
Sinorhizobium meliloti FcrX coordinates cell cycle and division during free-living growth and symbiosis by a ClpXP-dependent mechanism
In this study, a new essential player of cell cycle regulation has been characterized in the nitrogen fixing symbiont Sinorhizobium meliloti. During the nitrogen-fixing symbiosis between the alphaproteobacterium Sinorhizobium meliloti and the plant Medicago sativa (alfalfa), the interplay of molecular mechanisms governing cell cycle and bacteroid differentiation is a remarkable system that has still many details to be discovered. Here, we describe a bacterial cell cycle regulator, named FcrX, that controls two of the main essential players of cell cycle and bacteroid differentiation: the master regulator CtrA and the tubulin-like Z-ring component FtsZ. This essential factor is potentially participating with the degradosome complex driving the proteolysis of those two important regulators. Constitutive expression of FcrX during nodule development shows an increase of plant biomass, opening interesting paths in the amelioration of biological nitrogen fixation for a sustainable agriculture. More information : https://doi.org/10.1073/pnas.2412367122 Contact : Emanuele Biondi - emanuele.biondi@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 14, 2025 3:19 AM
|
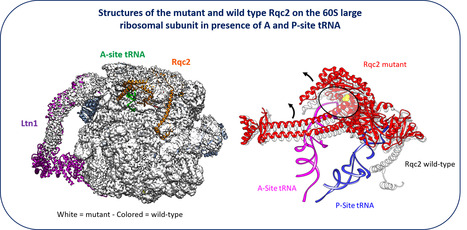
New facet of the Rqc2 protein part of a ribosome rescue pathway in Saccharomyces cerevisiae
The Ribosome Quality Complex protein 2 is a major player in peptide release from stalled ribosomes. Cells employ many quality control mechanisms during protein biosynthesis. Defects impairing messenger RNA decoding can cause ribosomes to stall, thereby distorting gene expression. Ribosome stalling results in ribosomes being sequestered in an inactive state on mRNA and represents a challenge for both ribosome homeostasis and cell survival. Eukaryotic cells prevent the accumulation of potentially toxic aberrant polypeptides and maintain ribosome availability by employing surveillance and clearance mechanisms, including the evolutionarily conserved ribosome-associated quality control complex (RQC). The rescue pathways dissociate the stalled ribosome, leading to the release of a 40S subunit attached to the mRNA and a 60S subunit still attached to a tRNA with the elongating peptide. RQC mediates the ubiquitination, extraction, and rapid degradation of the aberrant nascent peptide. In particular, Rqc2p recruits charged alanyl- and threonyl-tRNAs at the same position as an A-site tRNA, resulting in C-terminal extensions composed of alanine and threonine residues (CAT tails). These CAT tails may expose potential tunnel-embedded lysine residues for ubiquitination by the E3 ligase Ltn1. Using a genetic screen, we identified a mutant allele of RQC2 as involved in peptide release from stalled ribosomes. We characterized the mutant protein both biochemically and structurally, in collaboration with Reynald Gillet’s team (IGDR, Rennes University). We determining how the underlying point mutation reduced tRNA recruitment to the A site, thereby limiting CAT tail formation. These findings provide further mechanistic insight into the role of Rqc2p in ribosome rescue and the maintenance of ribosome homeostasis. More Information:https://pubmed.ncbi.nlm.nih.gov/40187343/ Contact: Céline Fabret celine.fabret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 7, 2025 5:04 AM
|
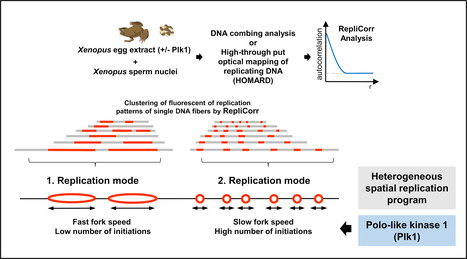
Yin-Yang during DNA replication
DNA duplication of the genetic material is a crucial step of cell proliferation. The study reveals that this step comprises two opposed strategies, coordinated by the enzyme Plk1. In vertebrates, DNA replication involves the activation of thousands of starting points for DNA copying, known as “origins of replication”. These points are irregularly scattered across the genome. However, the precise mechanisms controlling the coordinated activation of these starting points and the regulation of the rate at which DNA is copied remain poorly understood. Any dysfunction can lead to genetic abnormalities and promote the development of cancers.
The scientists studied DNA replication using cell-free extracts from the eggs of the amphibian Xenopus laevis. This model is quite similar to replication in human cells, and has made it possible to develop a new method for analyzing the distribution of replication starting points (initiations) and the speed of replication in individualized DNA molecules. Combining this analysis with kinetic models, the scientists discovered that DNA replication relies on two dynamic and opposing strategies:
A fast strategy: high replication speed, but with fewer starting points.
A slow strategy: a slower replication speed compensated by the activation of numerous starting points.
In this context, the scientists determined that the enzyme Polo-like kinase 1 (Plk1) played a key role in coordinating these two strategies. Plk1, frequently mutated in many cancers, regulates this balance, opening up new perspectives on the control of replication and its implications in oncology. (published by Paris-Saclay and CNRS Biologie Actu (https://www.insb.cnrs.fr/fr/cnrsinfo/yin-yang-dans-la-duplication-des-chromosomes ) More Information: https://doi.org/10.1093/nar/gkaf007 Contact: Arach Goldar arach.goldar@i2bc.paris-saclay.fr Kathrin Marheineke kathrin.marheineke@ijm.fr

|
Scooped by
I2BC Paris-Saclay
April 3, 2025 6:36 AM
|
Exploring the Interaction of Human α‐Synuclein with Polyethylene Nanoplastics: Insights from Computational Modeling and Experimental Corroboration
Can the presence of microplastics in the brain affect proteins? Plastics are now part of our daily lives because they are used in so many different ways. They are therefore a major source of pollution. Their chemical stability has consequently become a major concern for the environment and health, given their presence in all ecosystems. The penetration of plastic particles into living organisms through ingestion or inhalation has now been widely demonstrated. Micro- and nanoplastics are found throughout the human body, including in the brain, which raises the question of their potential toxicity. In a biological environment, the plastic particle does not remain naked but interacts with the surrounding molecules to form a biocorona. In this study, Tripathi et al. investigate the binding behavior of human α-synuclein (hαSn) with polyethylene (PE)-based plastics using molecular dynamics simulations and experimental methods. Their simulations show that (a) hαSn folds into a compact conformation to enhance intramolecular interactions, (b) non-oxidised PE nanoplastics facilitate the rapid adsorption of hαSn onto its surface with a change in the structural properties of hαSn, and (c) oxidised nanoplastics fail to capture hαSn. The experimental dynamic light scattering and adsorption isotherms are in good agreement with simulations. The observed formation of the plastic nanoparticle complex with hαSn can be proposed as a plausible pathogenic driving force in neuronal dysfunction and subsequent neurological damage. More information : https://doi.org/10.1021/acs.biomac.4c00918 Contact: Yves Boulard yves.boulard@i2bc.paris-saclay.fr
|

|
Scooped by
I2BC Paris-Saclay
December 8, 2025 5:52 AM
|
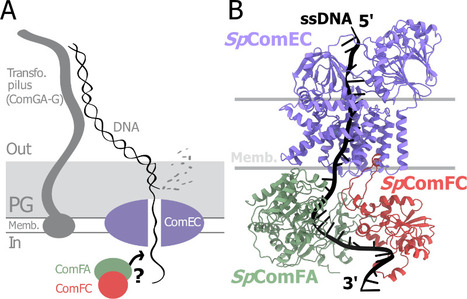
A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes.
From structural modeling to natural transformation in bacteria: identification of a key protein complex for DNA transport across the membrane of Firmicutes. Transformation is a key mechanism of horizontal gene transfer, central to bacterial adaptation. This evolutionarily conserved process allows bacteria to integrate exogenous genetic material into their genome, thereby facilitating, for example, the spread of antibiotic resistance.
In a study published in the journal PNAS, scientists from I2BC (CEA/CNRS/UPSaclay, Gif-sur-Yvette) and DRCM/IBFJ (CEA/UPSaclay/UP Cité, Fontenay-aux-Roses), in collaboration with a laboratory at CBI (CNRS/University of Toulouse), identified a protein complex involved in the transport of single-stranded DNA across the membrane during transformation. Using AlphaFold, they structurally modeled this three-protein complex, which is highly conserved in the phylum Firmicutes, in interaction with single-stranded DNA. The structural model allowed them to identify a possible path for DNA through a conserved channel in one of the three proteins, a transmembrane protein, and then along a groove formed by the other two proteins. This model was validated by a robust experimental strategy in the bacterium Streptococcus pneumoniae, by measuring the impact on transformation efficiency of disruptive mutations in several protein–protein and protein–DNA interfaces. The transmembrane channel was found to be conserved in a structural model in Helicobacter pylori, and its importance for transformation was also experimentally confirmed in this bacterium. This study sheds light on the molecular mechanisms of bacterial transformation and demonstrates the power of macromolecular structure prediction to generate molecular hypotheses and guide functional experiments. More information: https://doi.org/10.1073/pnas.2511180122 Contact : Jessica Andreani jessica.andreani@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 5, 2025 9:28 AM
|
Structural determinants of SlpA-mediated phage recognition in Clostridioides difficile
Study published in PLoS Pathogens of molecular features of phage receptor SlpA protein demonstrated the complexity of the interactions between human enteropathogen C. difficile and its phages The emergence of Clostridioides difficile as a leading cause of healthcare-associated nosocomial intestinal infections calls for novel therapeutic strategies, particularly in the context of antibiotic resistance and recurrent disease. Phage therapy is a promising approach, but its clinical application against C. difficile remains hindered by our lack of understanding of the factors driving host specificity. Indeed, it is crucial to understand how phages specifically interact with their host to be able to select the best candidates for cocktail preparation. The main surface layer protein SlpA is a key phage receptor, but the molecular details governing phage-receptor interactions remain unclear. By dissecting the structural features of SlpA required for phage infection through engineered SlpA isoforms and domain modifications, in collaboration with our Canadian collegues from Sherbrooke University, we reveal how specific regions of SlpA mediate phage adsorption and infection. These new insights into phage–receptor interactions will be instrumental in guiding the future engineering of broad-host-range therapeutic phages. More information : https://doi. org/10.1371/journal.ppat.1013724 Contact : Olga Soutourina olga.soutourina@i2bc.paris-saclay.fr https://www.i2bc.paris-saclay.fr/regulatory-rnas-in-clostridia/

|
Scooped by
I2BC Paris-Saclay
November 20, 2025 6:14 AM
|
Strict gut symbiont specificity in Coreoidea insects governed by interspecies competition
Insects choose their bacterial gut symbionts by creating a multifactorial selective environment that promotes competition among symbiont candidates and colonization by the single, best-adapted strain

|
Scooped by
I2BC Paris-Saclay
November 9, 2025 4:21 AM
|
Three stop codons to get over to flourish
Funded by the European Research Council (ERC synergy 2025), the project “3Stops2Go” leaps over the “three red lights” of premature stop codons to re-express critical protein and correct genetic diseases. The project, recently funded by an ERC Synergy Grant 2025, aims to target premature termination codon (PTC)mutations, which prematurely halt protein translation and are involved in about 11% of human genetic diseases.
By combining expertise in protist biology, RNA biology, and gene therapy, the consortium of four researchers (Leoš Valášek (Institute of Microbiology, Czech Academy of Sciences, Czech Republic), Julius Lukeš (Biology Centre, Czech Academy of Sciences, Czech Republic), Olivier Namy (Université Paris-Saclay, CEA, CNRS, France / I2BC, France), and Mark Osborn (University of Minnesota, USA)) aims to harness natural stop-codon bypass mechanisms to develop therapeutic tools (engineered tRNAs and readthrough inducers), test them in patient-derived cells and animal models, and ultimately pave the way toward clinical applications. More information: https://erc.europa.eu/news-events/news/synergy-grants-2025-examples-projects Contact: Olivier Namy olivier.namy@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 6, 2025 3:37 PM
|
New team in the Department of Genome Biology
Cécile Courret, CNRS Researcher and recipient of an ATIP-Avenir grant, has joined the Department of Genome Biology to establish her team ‘Intragenomic Conflict and Evolution’. Cécile's research focus on genetic conflicts and their impact on genome evolution. The basic principles of Mendelian inheritance state that in heterozygous individuals, two alleles have equal chances of being transmitted to the next generation. However, some genetic elements do not follow these rules. These so-called selfish genetic elements, such as transposons or meiotic drivers, bias inheritance in their own favor, often at the expense of the organism. Because they disrupt essential processes like meiosis, heterochromatin regulation, and cell division, selfish elements create a persistent conflict with the host genome. This conflict triggers an evolutionary “arms race,” in which genomes evolve defense mechanisms to counterbalance the harmful effects of these elements. Far from being rare exceptions, such conflicts are now recognized as a major force shaping genome structure and function. My research relies on the Drosophila model and combines genomic, molecular, and cytological approaches to investigate both how selfish elements perturb fundamental biological processes and how organisms respond to mitigate these disruptions. By studying systems where these conflicts are still active in natural populations, as well as the genomic signatures left by past events, Cécile's team aims to better understand how conflicts drive evolutionary innovation and shed light on the molecular basis of essential biological mechanisms. Contact: Cécile Courret, cecile.courret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 10, 2025 6:12 AM
|
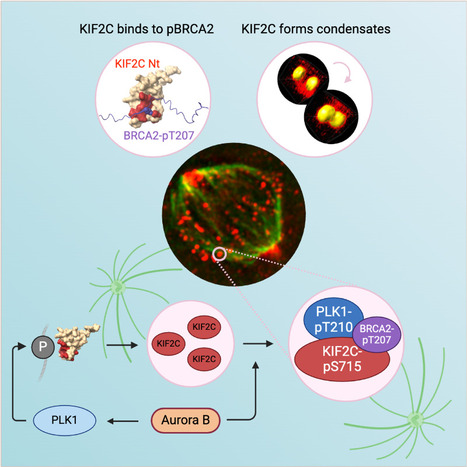
KIF2C condensation concentrates PLK1 and phosphorylated BRCA2 on kinetochore microtubules in mitosis
The microtubule depolymerase KIF2C forms membrane-less organelles at the kinetochore through its N-terminal phospho-peptide binding domain that interacts with BRCA2 and other phosphorylated targets in mitosis. During mitosis, the microtubule depolymerase KIF2C, the tumor suppressor BRCA2, and the kinase PLK1 contribute to the control of kinetochore-microtubule attachments. Both KIF2C and BRCA2 are phosphorylated by PLK1, and BRCA2 phosphorylated at T207 (BRCA2-pT207) serves as a docking site for PLK1. Reducing this interaction results in unstable microtubule-kinetochore attachments. Here we identified that KIF2C also directly interacts with BRCA2-pT207. Indeed, the N-terminal domain of KIF2C adopts a Tudor/PWWP/MBT fold that unexpectedly binds to phosphorylated motifs. Using an optogenetic platform, we found that KIF2C forms membrane-less organelles that assemble through interactions mediated by this phospho-binding domain. KIF2C condensation does not depend on BRCA2-pT207 but requires active Aurora B and PLK1 kinases. Moreover, it concentrates PLK1 and BRCA2-pT207 in an Aurora B-dependent manner. Finally, KIF2C depolymerase activity promotes the formation of KIF2C condensates, but strikingly, KIF2C condensates exclude tubulin: they are located on microtubules, especially at their extremities. Altogether, our results suggest that, during the attachment of kinetochores to microtubules, the assembly of KIF2C condensates amplifies PLK1 and KIF2C catalytic activities and spatially concentrates BRCA2-pT207 at the extremities of microtubules. We propose that this novel and highly regulated mechanism contributes to the control of microtubule-kinetochore attachments, chromosome alignment, and stability. More Information : https://academic.oup.com/nar/article/53/11/gkaf476/8160319 Contact: Sophie Zinn sophie.zinn@i2bc.paris-saclay.fr
Aujourd’hui, focus sur un nouvel équipement, et une expertise unique sur le territoire Paris-Saclay : Le tri de micro-organismes pathogènes par cytométrie en flux ! Pour rappel, cette technique permet de mesurer, évènement par évènement, des caractéristiques morphologiques (taille, complexité/granularité) et de fluorescence sur une population de cellules ou d’organites en suspension. Le tri par cytométrie en flux rend également possible la séparation physique et donc la purification de ces évènements suivant un ou plusieurs critères définis (morphologie, fluorescence) et à partir de populations hétérogènes. Nouvel équipement et expertise unique sur Paris-Saclay ? L’écoute et l’anticipation des besoins utilisateurs (dialogue, mais aussi via enquêtes de satisfaction et de besoins) sur la plateforme de cytométrie de l’I2BC (I2BC / Plateforme de cytométrie (I2BC - plateformes IMAGERIE-GIF, Gif-sur-Yvette, CNRS/Institut Joliot CEA/UPSaclay) ont souligné une nécessité de réaliser des approches en cytométrie en flux dans des espaces L2. L'accès à l'expertise et aux équipements pour réaliser de telles expériences est restreint dans le périmètre de l'Université Paris-Saclay, et un réel besoin de prestations en environnement L2 a été identifié. C’est pour répondre à ce besoin d’un accès « facile » à un cytomètre de tri « user-friendly » permettant à l’utilisateur d’être autonome et la nécessité de travailler en L2 pour les projets de tri de micro-organismes pathogènes que la plateforme de cytométrie s’est équipée d’un CytoFLEX SRT (Beckman) en espace L2 dans l’I2BC. Ce nouveau trieur « de paillasse », financé en partie par l’ERM Paris-Saclay, est installé sous PSM de type II dans un environnement L2 de l’I2BC, et dédié aux micro-organismes pathogènes. Sa configuration a été discutée pour assurer sa complémentarité et sa compatibilité avec l’analyseur existant. L’appareil est équipé de 2 lasers (488 nm et 561 nm), de 7 détecteurs de fluorescence (525/40 nm, 585/42 nm, 610/20 nm, 675/30 nm, 690/50 nm, 710/50 nm et 780/60 nm) et 2 détecteurs de taille/granularité. Cette configuration permettra de lire un grand nombre de fluorochromes endogènes (GFP, mCherry, dTomato…) et exogènes (nombreux AlexaFluor, nombreux marqueurs d’acides nucléiques, …) pour mieux identifier les cellules d’intérêt, même si elles sont rares. Une fois les cellules d’intérêt identifiées, elles pourront être triées dans des tubes (le trieur bénéficie de 4 voies de tri en sortie) ou sur des plaques de 6 à 96 puits. Les bactéries triées pourront être remises en culture ou utilisées pour des expériences de biologie moléculaire (RNAseQ, ChIPseQ, Hi-C, par exemple). Le CytoFLEX SRT permet de préparer une configuration de tri facile et automatisée, avec une mesure de contrôle qualité assurée par des billes de fluorescence. Tous ces paramètres en font un trieur facile à utiliser en autonomie. L’équipement sera donc en accès autonome après formation par les ingénieurs de la plateforme (Karine Madiona et Mickael Bourge). N’hésitez pas à contacter la plateforme pour mettre au point vos expériences de tri qui pourront débuter au cours du 1er trimestre 2025. -> Contact : Mickael Bourge et Karine Madiona (plt-cyto@i2bc.paris-saclay.fr) Plug In Labs Université Paris-Saclay : cliquer ICI La plateforme communique régulièrement via des FOCUS PLATEFORME. Envie de les relire ? I2BC / Plateforme de cytométrie (I2BC - plateformes IMAGERIE-GIF). La plateforme réalise environ 300 prestations par an pour divers groupes de recherche appartenant à différents organismes de tutelles ou de sociétés privées. Les publications du service traduisent les nombreuses collaborations développées avec les différents instituts de la communauté Paris-Saclay ainsi qu’avec d’autres partenaires tels que l’INRAe, l’INSERM, l’IRD, le CIRAD, des universités françaises et étrangères. Son expérience polyvalente et son expertise en sondes fluorescentes permettent d’adapter la cytométrie à des projets très divers issus de laboratoires publics et privés. Son partenariat avec SPS (Labex Saclay Plant Sciences) contribue à une activité importante dans le domaine de la biologie végétale. La plateforme de cytométrie en flux propose la mesure de fluorescence d'un ou plusieurs (> 10) fluorochromes simultanément, cellule par cellule. Cette technologie permet d'étudier: i) le dosage de la quantité d’ADN nucléaire en vue de l’étude de cycles cellulaires et d’endoréplication, ii) le dosage d’ADN à des fins de recherche en écologie et systématique, et d’amélioration des variétés (analyse de ploïdies), iii) le suivi de l’activité génique par l’expression d’un ou plusieurs gènes rapporteurs (tels que celui de la "Green Fluorescent Protein"-GFP et autres protéines fluorescentes), iv) des mesures d’activités métaboliques de la cellule (biosenseurs): dosage de calcium, pH, potentiel membranaire, poussées oxydatives, glutathion..., v) des analyses immunologiques, vi) le tri de cellules animales, de levures, de bactéries, de protoplastes et d’organites cellulaires. A propos de l’Institut de Biologie Intégrative de la Cellule (I2BC - UMR 9198). L’I2BC est une Unité Mixte de Recherche (CEA, CNRS, Université Paris-Saclay), accueillant une soixantaine d’équipes de recherche et hébergeant 17 plateformes technologiques, réparties en 6 pôles. 2025 est aussi une année clé pour l’I2BC : cette unité fête ses 10 ans cette année.
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
April 29, 2025 2:50 AM
|
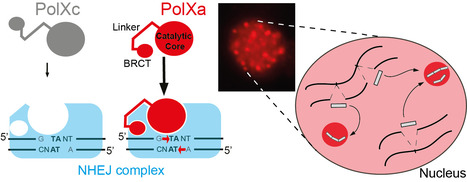
Paramecium PolX DNA polymerases keep the genome safe during programmed rearrangements
The expansion of PolX DNA polymerases in the ciliate Paramecium tetraurelia has favored the functional diversification of a subset of these enzymes, with enhanced nuclear localization in DNA repair foci during programmed DNA elimination. During its sexual cycle, Paramecium massively rearranges its genome by removing tens of thousands of internal eliminated sequences (IESs) from its developing somatic nucleus. This process involves the introduction of programmed DNA double-strand breaks (DSBs), followed by efficient DSB repair using the non-homologous end joining (NHEJ) pathway. In this system, DSB repair requires not only the core NHEJ factors Ku70/80 and Xrcc4/Lig4, but also additional enzymes that accurately process the 4-base 5′-overhangs of the broken DNA ends created at IES excision sites. In this study, we identify four orthologs of human DNA polymerase lambda in P. tetraurelia —PolXa, PolXb, PolXc, and PolXd— as key players in repairing IES excision junctions. Cells lacking these PolX enzymes accumulate genome-wide DNA damage, including unrepaired double-strand breaks, small deletions, and retained IESs. All four PolX proteins can process DNA ends, but PolXa and PolXb are specifically produced during programmed genome rearrangement and possess a unique internal linker region that enhances their nuclear localization. In contrast to PolXc, we show that PolXa concentrates in nuclear foci along with core NHEJ factors and a Dicer-like enzyme involved in producing IES-specific small RNAs that regulate IES excision. We propose that these foci are the sites where excised IESs are ligated into concatemers, which serve as templates for small RNA production, linking DNA repair to RNA-mediated regulation of genome dynamics. More information: https://academic.oup.com/nar/article/53/7/gkaf286/8113560 Contact: Mireille Bétermier mireille.betermier@i2bc-paris-saclay.fr, Julien Bischerour julien.bischerour@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 15, 2025 8:38 AM
|
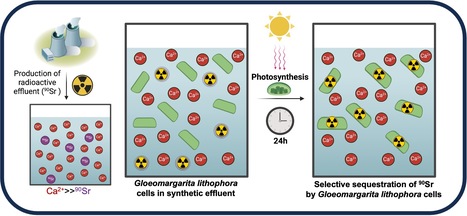
The cyanobacterium G. lithophora is a promising organism for effective bioremediation of waters contaminated with 90Sr
The unicellular cyanobacterium Gloeomargarita lithophora is able to stably sequester and withstand 90Sr and efficiently remove this hazardous anthropogenic radionuclide from aqueous solutions, including a synthetic nuclear effluent. Strontium (Sr) is an alkaline earth metal commonly occurring in nature. In aqueous environments, it prevails as Sr2+ ions form chemically similar to Ca2+. In most organisms, the non-selective Sr intake primarily comes from water and food. In human, it is mainly localized in bones and dental enamel and is involved in the control of bone formation. Radioactive isotopes such as 90Sr are artificially produced by nuclear fission. 90Sr, a b-emitter with a half-life of 28.8 years, is one of the most hazardous anthropogenic radionuclides. It has been massively released into the environment during nuclear weapons testing and nuclear reactor accidents, such as those at Chernobyl and Fukushima. It is also found in radioactive water effluents produced by nuclear power plants, requiring specific treatment before being discharged. In humans, its accumulation in bone tissues increases the risk of cancers. Current physico-chemical techniques for remediating 90Sr traces from effluents are costly and can exhibit a low selectivity for Sr over Ca. Therefore, there is an incentive to develop an alternative method of remediation. In this study, we demonstrate that the cyanobacterium Gloeomargarita lithophora can remove more than 90% of the 90Sr activity that could be found in a nuclear plant effluent within 24 hours. This process occurs through two steps: a first rapid and passive phase of 90Sr sorbtion to the cell surface, followed by an active phase of 90Sr accumulation within the cells, partially driven by photosynthesis. We also showed that this cyanobacterium is able to stably sequester and withstand 90Sr and to efficiently remove this hazardous radionuclide from aqueous solutions, including a synthetic nuclear effluent. These results highlight Gloeomargarita lithophora as a promising solution for effective bioremediation of water contaminated with 90Sr thereby safeguarding the well-being of our ecosystems.
More information : https://doi.org/10.1016/j.jhazmat.2025.138155 Contact : Corinne CASSIER-CHAUVAT Corinne.CASSIER-CHAUVAT@cea.fr

|
Scooped by
I2BC Paris-Saclay
April 11, 2025 5:59 AM
|
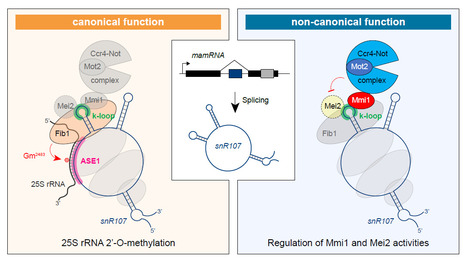
A bifunctional snoRNA guides rRNA 2’-O-methylation and scaffolds gametogenesis effectors
This study uncovers a fission yeast small nucleolar RNA (snoRNA) that guides ribosomal RNA 2’- O-methylation and modulates the activities of RNA-binding proteins involved in gametogenesis, expanding our vision of the non-canonical functions exerted by snoRNAs. Small nucleolar RNAs are non-coding transcripts that guide chemical modifications of RNA substrates and modulate gene expression at the epigenetic and post-transcriptional levels. However, the extent of their regulatory potential and the underlying molecular mechanisms remain poorly understood. In a collaborative work with the I2BC B3S department and NGS facility, the epiRNA-Seq facility in Nancy and the Palancade lab at the Institut Jacques Monod, we have identified a conserved, previously unannotated intronic C/D-box snoRNA, termed snR107, hosted in the fission yeast long non-coding RNA mamRNA and carrying two independent cellular functions. On the one hand, snR107 guides site-specific 25S rRNA 2’-O-methylation and promotes pre-rRNA processing and 60S subunit biogenesis. On the other hand, snR107 associates with the gametogenic RNA-binding proteins Mmi1 and Mei2, mediating their reciprocal inhibition and restricting meiotic gene expression during sexual differentiation. Both functions require distinct cis-motifs within snR107, including a conserved 2’-O-methylation guiding sequence. Together, our results position snR107 as a dual regulator of rRNA modification and gametogenesis effectors, expanding our vision on the non-canonical functions exerted by snoRNAs in cell fate decisions. More Information: https://www.nature.com/articles/s41467-025-58664-y Contact: Mathieu Rougemaille mathieu.rougemaille@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 3, 2025 8:51 AM
|
A monomer–dimer switch modulates the activity of plant adenosine kinase
Novel regulation of adenosine kinase activity in plants Adenosine undergoes ATP-dependent phosphorylation catalyzed by adenosine kinase (ADK). In plants, ADK also phosphorylates cytokinin ribosides, transport forms of the hormone. Here, we investigated the substrate preferences, oligomeric states, and structures of ADKs from moss (Physcomitrella patens) and maize (Zea mays) alongside metabolomic and phenotypic analyses. We showed that dexamethasone-inducible ZmADK overexpressor lines in Arabidopsis can benefit from a higher number of lateral roots and larger root areas under nitrogen starvation. We discovered that maize and moss enzymes can form dimers upon increasing protein concentration, setting them apart from the monomeric human and protozoal ADKs. Structural and kinetic analyses revealed a catalytically inactive unique dimer. Within the dimer, both active sites are mutually blocked. The activity of moss ADKs, exhibiting a higher propensity to dimerize, was 10-fold lower compared with maize ADKs. Two monomeric structures in a ternary complex highlight the characteristic transition from an open to a closed state upon substrate binding. This suggests that the oligomeric state switch can modulate the activity of moss ADKs and probably other plant ADKs. Moreover, dimer association represents a novel negative feedback mechanism, helping to maintain steady levels of adenosine and AMP. More information: https://academic.oup.com/jxb/advance-article/doi/10.1093/jxb/eraf094/8068880 Contact: Solange Morera solange.morera@i2bc.paris-saclay.fr
|





 Your new post is loading...
Your new post is loading...