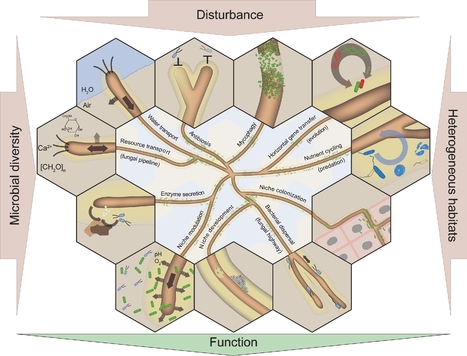
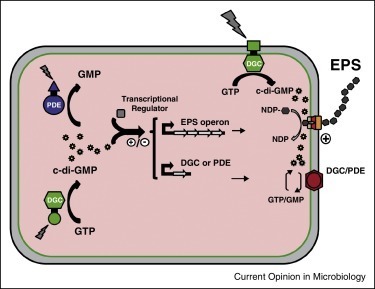
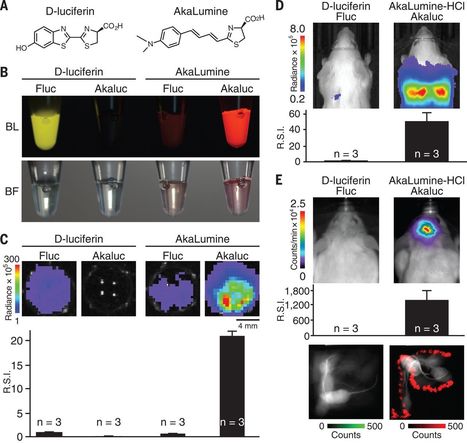
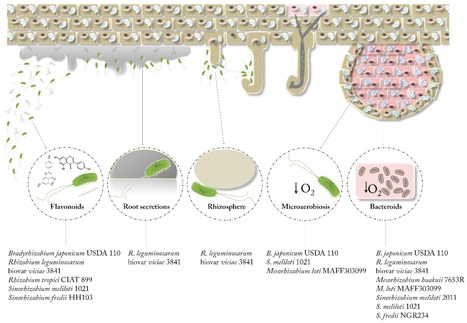
Fungi and bacteria are found living together in a wide variety of environments. Their interactions are significant drivers of many ecosystem functions and are important for the health of plants and animals. A large number of fungal and bacterial families are engaged in complex interactions that lead to critical behavioural shifts of the microorganisms ranging from mutualism to pathogenicity. The importance of bacterial-fungal interactions (BFI) in environmental science, medicine and biotechnology has led to the emergence of a dynamic and multidisciplinary research field that combines highly diverse approaches including molecular biology, genomics, geochemistry, chemical and microbial ecology, biophysics and ecological modelling. In this review, we discuss most recent advances that underscore the roles of BFI across relevant habitats and ecosystems. A particular focus is placed on the understanding of BFI within complex microbial communities and in regards of the metaorganism concept. We also discuss recent discoveries that clarify the (molecular) mechanisms involved in bacterial-fungal relationships, and the contribution of new technologies to decipher generic principles of BFI in terms of physical associations and molecular dialogues. Finally, we discuss future directions for researches in order to catalyse a synergy within the BFI research area and to resolve outstanding questions.
Follow, research and publish the best content
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
|
|