Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:32 AM
|
Translational regulation offers a powerful biological control axis with the potential to enable programmable control over synthetic mRNAs. Here, we introduce inducible Deaminases Acting on RNA (iDARs): deaminase domains (DDs) with conditional RNA-editing activities. Using a domain-insertion strategy, we designed autoinhibited enzymes that can be converted into active RNA editors in response to triggers based on small molecules (chemiDARs), intracellular antigens (antiDARs), protease cleavage (lysiDAR), and optical excitation (optiDAR). Coupling these domains with novel stop codon containing RNA substrates enabled conditional protein translation or transcript degradation. Mutational tuning of inositol hexaphosphate (IP6)-binding pockets produced tightly regulated deaminases with minimal basal activity, facilitating dose-dependent readthrough translation in response to low-nanomolar drug concentration, with dynamic ranges exceeding 100-fold. By encoding iDARs alongside their substrates, we developed 'self-editing' polycistronic transcripts capable of directing translation of encoded proteins in a trigger-dependent manner following delivery to cells as in vitro transcribed mRNAs. Overall, iDARs provide a generalizable framework for generating controllable deaminases, enabling the design of post-transcriptional circuits that link biochemical sensing to readouts based on de novo translation or mRNA decay.

|
Scooped by
mhryu@live.com
Today, 1:06 AM
|
Selective encapsulation of target enzymes is an increasingly well studied field with a host of potential applications for biotechnology. Natively, many bacteria utilize bacterial microcompartments (BMC) for enzyme encapsulation to enhance catalysis. BMCs are protein shells that enable selective localization of targeted metabolic enzymes and may improve catalytic rates by co-localizing pathway enzymes and/or serve to sequester toxic or volatile intermediates. The microcompartment shell of Haliangium ochraceum (HO) is a notable BMC chassis because of its modularity and versatility; it is easily expressed and assembled outside its native host and can accept a wide array of cargo. Recently, it was demonstrated that assembly of HO BMC shells can be easily achieved in vitro. Following up on our previous work on in vivo assembly of HO-BMCs with triose phosphate isomerase (TPI) as model enzyme cargo, here we have demonstrated the advantages of in vitro assembly (IVA) for targeted enzyme encapsulation. We achieved variable loading of BMC shells with targeted amounts of TPI and demonstrated enhanced thermal stability of encapsulated TPI versus free TPI up to 62°C.

|
Scooped by
mhryu@live.com
Today, 12:59 AM
|
Methanol, a promising one-carbon (C1) feedstock for biofuels, faces challenges in bioconversion due to its cellular toxicity. This review summarizes recent advances in methanol-based biosynthesis of biofuels, such as short-chain alcohols, fatty acid derivatives, and terpenoids, in both native and synthetic methylotrophs. We also discuss the mechanisms of methanol cytotoxicity and systematically examine engineering strategies to enhance methanol utilization and tolerance, including metabolic pathway rewiring, compartmentalization, and adaptive evolution. Finally, we highlight that integrating systems biology and synthetic biology can pave the way toward sustainable methanol-based biomanufacturing.

|
Scooped by
mhryu@live.com
Today, 12:44 AM
|
Indole-3-acetic acid (IAA) is a tryptophan-derived gut microbial metabolite with reported anti-inflammatory activities, but the organisms and anaerobic pathways that support robust production remain unclear. Screening 206 human gut bacterial isolates by LC-MS revealed that IAA production is rare: only five strains exceeded the limit of quantitation, and high-capacity production was confined to the acetogens Blautia hydrogenotrophica and Intestinibacter bartlettii. Across growth conditions, IAA was a minor product that rose alongside carbohydrate-sensitive, OFOR-linked catabolism of multiple amino acids, generating abundant branched-chain and aromatic organic acids. In gnotobiotic mice mono-colonized with I. bartlettii, these metabolites were produced in vivo but showed distinct host handling, with branched-chain fatty acids largely extracted between portal and peripheral plasma, whereas aromatic acids and their glycine conjugates appeared in plasma and urine. Genomic analyses and heterologous enzyme assays identified expanded repertoires of 2-oxoacid:ferredoxin oxidoreductases (OFORs) with activities spanning pyruvate/oxaloacetate, branched-chain, and aromatic 2-oxoacids, including indolepyruvate conversion to indoleacetyl-CoA, a putative intermediate en route to IAA. Finally, position-specific 13C tracing showed that CO2 released during amino acid oxidation is reassimilated into acetate via reductive acetogenesis, indicating that gut acetogens can maintain redox balance without fermenting partner strains. Together, these findings show that high IAA output is restricted to select gut acetogens and linked to a broader OFOR-driven anaerobic metabolism that generates additional metabolites that are absorbed by the host.

|
Scooped by
mhryu@live.com
January 29, 11:42 PM
|
Droplet digital (dd) clustered regularly interspaced short palindromic repeats (CRISPR) integrates the high sequence specificity of CRISPR-based nucleic acid detection with the absolute quantification capability of digital droplet microfluidics, offering high sensitivity, precision, and scalability. By partitioning samples into thousands to millions of picoliter microdroplets, ddCRISPR enables single-molecule resolution and minimizes background interference. This review summarizes the principles of droplet generation, manipulation, and detection in ddCRISPR platforms, as well as recent advances in amplification-based and amplification-free detection strategies. Representative applications are highlighted for viral, bacterial, and other DNA/RNA biomarker detection. Current challenges, including workflow automation, droplet stability, multiplexing, and assay portability, are discussed alongside future perspectives such as artificial intelligence (AI)-assisted analysis, point-of-care integration, and high-throughput multiplexed detection. These insights aim to guide the translation of ddCRISPR technologies from laboratory research to robust, scalable, and accessible diagnostic solutions.

|
Scooped by
mhryu@live.com
January 29, 11:27 PM
|
Artificial biomolecular condensates have emerged as powerful tools to control cellular behaviors. Here we introduce a method to build artificial condensates within living mammalian cells through the design of modular RNA motifs formed by a single, short strand of RNA. These condensates emerge spontaneously, creating RNA-rich compartments that remain separated from their surrounding environment. The RNA sequences include stem-loop domains that fold as the RNA is transcribed, and then condense in the nucleus and cytoplasm through loop-loop interactions. These sequences can be optimized and diversified, enabling the generation of distinct, non-mixing condensate populations and the programmable control of their subcellular localization. The RNA motifs can also be modified to recruit small molecules, proteins, and RNA molecules in a sequence-specific manner to the RNA-rich phase. By introducing additional RNAs that link two distinct types of condensates, we can create droplets with multiple subcompartments, whose organization can be controlled by tuning the stoichiometry of different RNA sequences. These artificial condensates provide a versatile platform for studying and manipulating molecular functions inside living cells.

|
Scooped by
mhryu@live.com
January 29, 4:55 PM
|
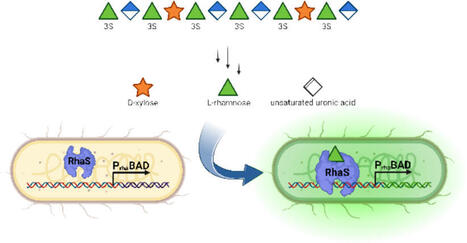
Marine macroalgae, particularly their complex polysaccharides, are an untapped renewable source of high-quality monosaccharides and related building blocks. To utilize this feedstock for industrial applications, the enzymatic depolymerization by marine microorganisms has been shown to be effective. A prime example is the common green alga Ulva, with its storage polysaccharide ulvan, which contains high quantities of L-rhamnose and D-glucuronic acid. As suitable high-throughput methods for analyzing the enzymatic degradation of complex polysaccharides are still lacking, a transcription factor–based biosensor is described here that utilizes the PrhaBAD promoter native to E. coli, which is specific for L-rhamnose. This biosensor exhibited a linear response, enabling the quantification of L-rhamnose within a concentration range of 10–1000 µM. The introduction of a T7 stem-loop improved the performance, and various fluorescent reporter genes were studied. The optimized system was then used to evaluate various stages of the ulvan degradation cascade in terms of L-rhamnose release, confirming its applicability to complex sugar mixtures. A detectable fluorescence signal was only generated when all the necessary enzymes for breaking down the polymer into undecorated monosaccharides were present, highlighting the biosensor’s specificity. The application of this method to the degradation of Ulva sp. biomass samples of various origins was also successfully demonstrated. This establishes the biosensor as a promising method for further high-throughput investigations.

|
Scooped by
mhryu@live.com
January 29, 4:41 PM
|
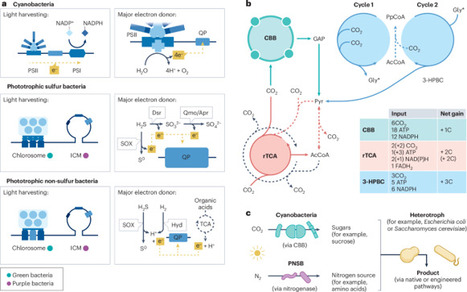
Phototrophic bacteria use solar energy to support metabolism and biochemical synthesis and are of increasing interest as systems for industrial chemical manufacture. Biosynthesis using phototrophic bacteria can use various renewable organic and inorganic carbon sources to drive chemical production, without an exogenous supply of refined organic carbon required for traditional dark fermentation used in industrial biotechnology. The potential to use solar energy to convert CO2 into useful products or upcycle diverse waste streams opens avenues for chemical manufacturing decoupled from fossil resource depletion, potentially with a smaller environmental footprint than other biotechnological routes. Despite this potential, the commercial application of phototrophic bacteria for this purpose is currently limited. In this Review, we discuss the basis for solar chemical bioprocesses in bacteria, emerging tools to engineer phototrophy for bioproduction and give examples of bulk chemical and high-value products synthesized in these species. Finally, we discuss the outlook for this nascent field in the context of chemical synthesis through engineering biology and outline the further progress required to realize the potential of light-powered microbial cell factories for future sustainable industrial synthesis. Phototrophic bacteria could be used for chemical manufacturing from various carbon sources. This Review discusses the pathways, engineering and potential application of solar chemical biosynthesis.

|
Scooped by
mhryu@live.com
January 29, 4:21 PM
|
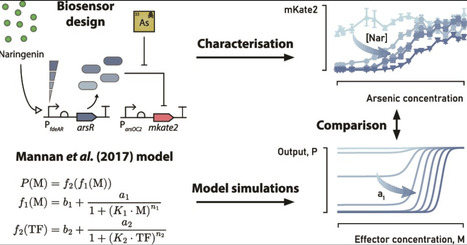
Whole-cell biosensors detecting the heavy metal arsenic have been widely studied for their potential in environmental monitoring. And while inducible biosensors have been shown to be an effective tool to tune the operational range, a thoroughly characterized inducible biosensor is currently lacking. Here, we present an E. coli biosensor for arsenic in which the transcription factor (TF) gene arsR is inducible by naringenin, a plant-derived secondary metabolite. Increasing the naringenin concentration reduced the basal output while increasing both the dynamic range and sensing threshold of the biosensor dose-response curve, but the operational range appeared constrained by a fixed upper limit. Comparison with a previously published phenomenological model revealed good overall agreement between experimental data and model predictions, except for the behavior of the maximum output and threshold. This work expands the biosensor toolbox with a profoundly characterized arsenic biosensor and raises a potential practical limit to dose-response curve engineering by tuning TF expression alone.

|
Scooped by
mhryu@live.com
January 29, 4:10 PM
|
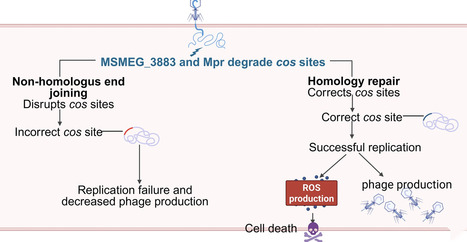
In the ongoing arms race with phages, bacteria have evolved diverse defense systems, such as CRISPR–Cas and restriction–modification systems. The DNA double-strand break repair system represents a core mechanism for maintaining genomic integrity and is vital for cell survival. However, it remains unknown whether and how these repair systems contribute to phage resistance. This study systematically investigates the role of the non-homologous end joining (NHEJ) during phage infection in Mycobacterium smegmatis. We found that NHEJ deficiency compromises host resistance to phage SWU1, as evidenced by increased plaque counts and reduced bacterial survival. Mechanistically, phages exploit host NHEJ for genomic repair; however, the error-prone nature of NHEJ leads to imperfect repair at phage cos sites, thereby blocking replication. The host modulates the balance between NHEJ and homologous recombination (HR) to control repair fidelity: NHEJ loss shifts the balance toward high-fidelity HR, which in turn promotes phage survival. Furthermore, NHEJ deficiency exacerbates infection-induced oxidative stress, leading to a compromise in bacterial viability. Our findings reveal the multifaceted functions of NHEJ in mycobacterium–phage interactions and provide new insights into how DNA repair systems shape antiphage defense and coevolution.

|
Scooped by
mhryu@live.com
January 29, 3:48 PM
|
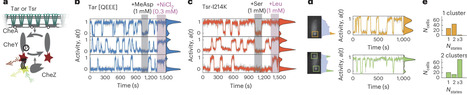
Cooperative interactions within large protein assemblies are crucial for cellular information processing. However, direct observations of cooperative transitions have been limited to compact molecular assemblies. Here we report the in vivo measurements of spontaneous discrete-level transitions in the activity of an entire E. coli chemosensory array—an extensive membrane-associated assembly comprising thousands of molecules. Finite-size scaling analysis of the temporal statistics reveals nearest-neighbor coupling strengths within 3% of the Ising phase transition, indicating that chemosensory arrays are poised at criticality. We also show how E. coli exploits both static and dynamic disorder, arising from chemoreceptor mixing and sensory adaptation, respectively, to temper the near-critical dynamics. This tempering eliminates detrimental slowing of response while retaining substantial signal gain as well as an ability to modulate physiologically relevant signal noise. These results identify near-critical cooperativity as a design principle for balancing the inherent trade-off between response amplitude and response speed in higher-order signalling assemblies. Many biological systems appear to organize their dynamics close to a critical point. Now it is shown that the protein array mediating Escherichia coli chemosensing is near-critical, enabling large signal amplification without compromising response speeds.

|
Scooped by
mhryu@live.com
January 29, 9:48 AM
|
Specialized metabolites encoded by biosynthetic gene clusters (BGCs) in the oral microbiome remain largely unexplored in the context of oral health and disease. Previous genome-centric surveys have identified hundreds of uncharacterized BGCs in the oral cavity associated with health and disease, but these studies relied on reference genomes and did not capture strain-level variation or the native distribution of BGCs. Here, we assembled three independently sourced metagenomic datasets from healthy and dental caries samples, extracted BGCs, and quantified their metagenomic abundance and transcriptional activity. We found that aryl polyene, ribosomally synthesized and post-translationally modified peptide (RiPP), and nonribosomal peptide (NRPS) encoding BGCs were the most prominent BGCs identified across the three metagenomic datasets. We grouped the identified BGCs into homology-based gene cluster families (GCFs) and found that specific GCFs were consistently associated with either health or caries across diverse taxa, suggesting that some specialized metabolites may perform conserved ecological functions. Conversely, other BGCs showed more restricted taxonomic distributions and were linked to disease-associated taxa, such as Propionibacterium acidifaciens, suggesting niche-specific biosynthetic capacities within the oral environment. Applying elastic-net regression to the metatranscriptomic dataset further identified a subset of 51 BGCs out > 3 000 that distinguished healthy from caries samples, reinforcing the discriminatory power of BGC expression patterns. Together, these results demonstrate that BGCs provide functional resolution beyond taxonomic profiling and that BGC expression, rather than genomic presence alone, differentiates oral microbial community states. This underscores the relevance of specialized metabolism to oral health and supports the use of BGC-centric analyses to interrogate microbial interactions underlying community stability and disease-associated shifts.

|
Scooped by
mhryu@live.com
January 29, 12:54 AM
|
Metagenomic sequencing has revolutionised the field of microbial ecology, as it has led to cultivation-independent exploration of complicated microbial communities. The assembly of metagenome-assembled genomes has provided genome-scale information about uncultivated microorganisms, but issues such as sequencing errors, fragmented assemblies, residual redundancy, uneven coverage, recovery of low-abundance taxa, and highly diversified taxa continue to impair the quality of these genomes. The latest achievements in artificial intelligence, particularly in machine learning and deep learning, have played a significant role in overcoming these limitations by enhancing quality control, error correction, assembly, binning, refinement, and annotation procedures. It is demonstrated that representation learning and graph-based binning methods have high strain-level resolution and can reduce contamination in complex microbial communities, whereas artificial intelligence-based assemblers and polishing tools improve base-level precision and assembly contiguity. This review synthesizes traditional and artificial intelligence-based workflows involved in the reconstruction of metagenome-assembled genomes, encompassing quality control, assembly, binning, refinement, and annotation, as well as quantitative benchmarking of significant artificial intelligence-based pipelines. As future directions, the focus on emerging trends, such as explainable artificial intelligence, federated learning, cloud-native scalable pipelines, multimodal and multi-omics integration, and large language model-based annotation, is covered. In general, the incorporation of artificial intelligence represents a paradigm shift in the reconstruction of metagenome-assembled genomes, allowing for a more relevant, scalable, and biologically informative search of the microbial dark matter in various ecosystems.
|

|
Scooped by
mhryu@live.com
Today, 1:07 AM
|
Volatile organic compounds (VOC) emitted by soil bacteria influence interactions with other soil microbes and with plant roots. While their potential as plant-growth promoters is well recognized, their role in promoting plant resilience to abiotic stress and the underlying molecular mechanisms remains poorly understood. Here, we investigate the role of Pseudomonas VOCs in enhancing plant resilience to drought stress. Arabidopsis thaliana plants were exposed to VOCs emitted by Pseudomonas strains under both control and osmotic-stress conditions. VOC exposure generally enhanced plant growth, and this effect was even more pronounced under both drought and salt stress. Transcriptomic analysis revealed that VOC exposure modulates key stress-responsive pathways, including those related to abscisic acid biosynthesis and signalling, sugar transport, iron uptake, aliphatic glucosinolate biosynthesis, and plant defences. Using Arabidopsis mutants, we identified abscisic acid and aliphatic glucosinolates as important components in mediating the plant response to VOCs. SWEET11/12 sugar transporters and ABA signaling genes were downregulated by VOCs exposure, in order to allow for a positive regulation of lateral root numbers (in case of SWEET genes) and plant growth in general under drought stress. In summary, using metabolomics, transcriptomics and functional analysis, we showed a negative cross-talk between the effects of VOCs on plant growth and glucosinolate production, whereas a positive interaction was observed between the biosynthesis of coumarins and VOCs. Notably, VOCs also improved drought tolerance in soil-grown Brassica oleracea plants. We showed that VOC treatment altered the root-associated microbiome under drought, leading to a community composition more similar to that of well-watered plants. Our results show that Pseudomonas emitted VOCs can promote plant growth under drought conditions, linked to root transcriptional reprogramming and direct or indirect microbiome modulation.

|
Scooped by
mhryu@live.com
Today, 1:01 AM
|
CRISPR interference (CRISPRi) has emerged as a versatile approach for targeted gene repression in many organisms, including microbes and bacteria, due to the simple design of sequence-specific transcriptional silencing of gene expression. However, the strain-specific effects on repression efficiency and the host when translating a CRISPRi system from a laboratory strain to non-model strains are not well understood, yet they can present important limitations to its use. Here, we investigated the repression efficiency and toxicity of three CRISPRi systems (one dCas9 and two dCas12a variants) across four different E. coli strains, including a laboratory K-12 strain (MG1655) and three non-model strains that are clinical isolates (probiotic Nissle 1917, uropathogenic CFT073, and uropathogenic UMN026). We evaluated the repression in each strain using sets of guide RNAs (gRNAs) targeting along the gene sequence and assayed cytotoxicity of expressing each dCas protein. Growth toxicity from expression of the different dCas proteins notably differed and showed high variation between some host strains. We also observed variable repression among the strains and notably poorer repression in multiple clinical strains. Therefore, we developed a dual gRNA CRISPRi system for enhanced gene silencing among the strains, which achieved up to 824-fold repression in CFT073. The results demonstrate that strain-specific design considerations can arise when a CRISPRi genetic system is transferred to a closely related bacterial strain. These findings provide insight into the relationships between criteria used for CRISPRi genetic design and in vivo activity across non-model E. coli strains, providing guidelines for diverse applications of these tools.

|
Scooped by
mhryu@live.com
Today, 12:46 AM
|
Agriculture is under pressure to provide food for a growing population and the feedstock required to drive the bioeconomy. Methods to breed and genetically modify plants are inadequate to keep pace. When engineering crops, traits are painstakingly introduced into plants one-at-a-time, combine unpredictably, and are continuously expressed. Synthetic biology is changing these paradigms with new genome construction tools, computer aided design (CAD), and artificial intelligence (AI). “Smart plants” contain circuits that respond to environmental change, alter morphology, or respond to threats. Further, the plant and associated microbes (fungi, bacteria, archaea) are now being viewed by genetic engineers as a holistic system. Historically, plant health has been enhanced by many natural and laboratory-evolved soil microbes marketed to enhance growth or provide nutrients, or pest/stress resistance. Synthetic biology has expanded the number of species that can be engineered, increased the complexity of engineered functions, controlled environmental release, and can assemble stable consortia. New CAD tools will manage genetic engineering projects spanning multiple plant genomes (nucleus, chloroplast, mitochondrion) and the thousands of genomes of associated bacteria/fungi. This review covers advanced genetic engineering techniques to drive the next agricultural revolution, as well as push plant engineering into new realms for manufacturing, infrastructure, sensing, and remediation.

|
Scooped by
mhryu@live.com
January 29, 11:47 PM
|
Fold switching, where a protein region interconverts between entirely distinct three-dimensional structures, is emerging as vital for certain protein functions. Here, we report a remarkable example in the F7 pyocin, a phage tail-like bactericidal nanomachine. Cryogenic electron microscopy and tomography reveal that a 163-residue segment of the central tail fiber undergoes a dramatic transition—from a trimeric α-helical coiled-coil to a triangular β-prism—upon binding to the bacterial cell surface. This massive fold switch remodels the tail tip, ejects the internal tape measure protein, and drives membrane puncture. Site-directed mutations that selectively destabilize the β-prism conformation completely abolish bactericidal activity without impairing particle assembly, implying that the energy released during this transition powers penetration. AlphaFold-based analyses further predict similar large-scale coiled-coil to β-prism switches in diverse non-contractile phage tails. This discovery reveals a sophisticated, ATP-independent strategy for microbial warfare and opens exciting possibilities for engineering next-generation bacteriocins to combat multidrug-resistant pathogens.

|
Scooped by
mhryu@live.com
January 29, 11:32 PM
|
Many soil protists are bacterivores, yet how protist predation reshapes bacterial metabolic interactions and functions remains poorly understood. Here, we combine global soil samples with microbial metabolic simulations, along with soil microcosm-pot validations, to investigate the influence of protists on bacterial metabolic interactions. Across 3,785 metabolic simulations spanning 757 soils, increased protists predicted higher bacterial metabolic interaction potential and cross-feeding but lower metabolic resource overlap and competition. These patterns were confirmed using an independent rhizosphere dataset and metagenomic analysis. Protist predation selected bacterial communities containing GC-rich genomes, acid-carbon-preferring taxa, and enhanced metabolite exchange. Additionally, exposing a synthetic community (SynCom) to protist predation elevated the expression of bacterial genes associated with plant growth-promoting functions. Consistently, microcosm- and pot-based experiments showed that protist addition increased bacterial cross-feeding over time and improved plant performance. Together, we establish a scalable framework to evaluate protist-driven bacterial cooperation and function to guide rational rhizosphere microbiome engineering.

|
Scooped by
mhryu@live.com
January 29, 10:41 PM
|
Microbial metabolites play a critical role in regulating ecosystems, including the human body and its microbiota. However, understanding the physiologically relevant role of these molecules, especially through liquid chromatography tandem mass spectrometry (LC-MS/MS)-based untargeted metabolomics, poses significant challenges and often requires manual parsing of a large amount of literature, databases, and webpages. To address this gap, we established the Collaborative Microbial Metabolite Center knowledgebase (CMMC-KB), a platform that fosters collaborative efforts within the scientific community to curate knowledge about microbial metabolites. The CMMC-KB aims to collect comprehensive information about microbial molecules originating from microbial biosynthesis, drug metabolism, exposure-related molecules, food, host-derived molecules, and, whenever available, their known activities. Molecules from other sources, including host-produced, dietary, and pharmaceutical compounds, are also included. By enabling direct integration of this knowledgebase with downstream analytical tools, including molecular networking, we can deepen insights into microbiota and their metabolites, ultimately advancing our understanding of microbial ecosystems.

|
Scooped by
mhryu@live.com
January 29, 4:51 PM
|
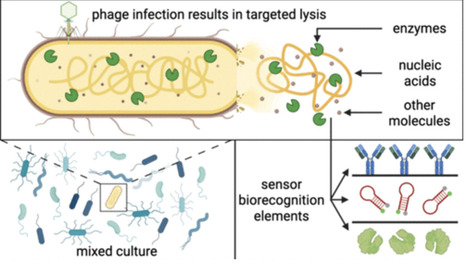
Cell lysis to release intracellular targets is a vital step in many bacterial sensing platforms and is often achieved through chemical or physical approaches. However, these conventional methods can have certain limitations such as cost, required equipment, safety, or risk of target damage. Cell lysis induced by bacteriophages, which are bacteria-infecting viruses, has some notable advantages, including safety and the self-amplifying properties of phage. Bacteriophages also induce species-selective infection, enabling the targeted lysis of a specific bacterial species in mixed cultures. Despite this, bacteriophage-induced lysis has to date been relatively poorly adopted in the bacterial biosensing field. In this Perspective, we outline the potential benefits of bacteriophage lysis in biosensors, while also exploring the reasons that it has not been more widely adopted. We also identify future research directions to facilitate increased incorporation of bacteriophages into bacterial detection platforms, including improving the characterization, availability, and stability of phage strains.

|
Scooped by
mhryu@live.com
January 29, 4:36 PM
|
Engineered bacteria are emerging as a transformative class of cancer therapeutics. Recent advances in synthetic biology have expanded the genetic circuit toolbox, enabling the programmable control of attenuation, payload release, and immunomodulation. These developments have transformed bacteria from simple, colonizing agents into a versatile chassis for complex therapeutic functions. In this review, we examine recent circuit-based strategies for enhancing tumor specificity, regulating therapeutic delivery and engaging the host immune system, with emphasis on programming spatiotemporal control and consortia behavior. We consider current barriers to clinical translational and discuss how rational engineering can guide the next generation of microbial therapeutics.

|
Scooped by
mhryu@live.com
January 29, 4:15 PM
|
Pseudomonas aeruginosa is a major cause of healthcare-associated infections and a significant threat to global health, primarily due to its ability to form biofilms that protect it from host immune responses and block antibiotic efficacy. While bacteriophages are emerging as potential antimicrobial agents, their effectiveness is often limited by these bacterial biofilms. This study aimed to enhance the biofilm-disrupting capabilities of phages through genetic engineering. First, we validated the in vitro biofilm-inhibitory effects of two enzymes: the quorum-quenching lactonase (Aiia) and a phage-derived depolymerase (DP). To demonstrate their potential, we then used CRISPR-Cas9 to engineer the P. aeruginosa phage PaGZ-1 to express these biofilm-disrupting genes. The resulting engineered phages demonstrated superior inhibition of biofilm formation compared to the wild-type phage. Notably, the PaGZ-1-Aiia variant showed significant promise in both inhibiting biofilm formation and disrupting established biofilms. Our findings provide a straightforward method for introducing exogenous genes into non-model P. aeruginosa phage genomes, offering a novel and potentially effective strategy for combating drug-resistant, biofilm-forming infections.

|
Scooped by
mhryu@live.com
January 29, 3:54 PM
|
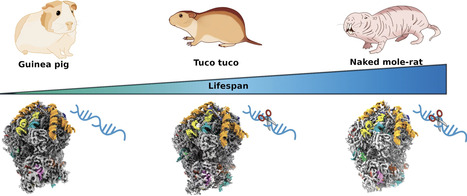
Ribosomes are central to protein synthesis in all organisms. In mammals, the ribosome functional core is highly conserved. Remarkably, two rodent species, the naked mole-rat (NMR) and tuco-tuco, display fragmented 28S ribosomal RNA (rRNA), coupled with high translational fidelity and long lifespan. The unusual ribosomal architecture in the NMR and tuco-tuco has been speculated to be linked to high translational fidelity. Here, we show, by single-particle cryo-electron microscopy, that despite the fragmentation of their rRNA, NMR and tuco-tuco ribosomes retain their core functional architecture. Compared to ribosomes of the guinea pig, a phylogenetically related rodent without 28S rRNA fragmentation, ribosomes of NMR and tuco-tuco exhibit poorly resolved density for certain expansion segments. In contrast, the structure of the guinea pig ribosome shows high similarity to the human ribosome. Enhanced translational fidelity in the NMR and tuco-tuco may stem from subtle, allosteric effects in dynamics, linked to rRNA fragmentation.

|
Scooped by
mhryu@live.com
January 29, 1:17 PM
|
Protein–carbohydrate interactions play an important role in many biological processes and functions, like inflammation, signal transduction, and cell adhesion. In our work, we will study non-covalent carbohydrate binding sites. In this paper, we aim to build a deep-learning model to predict non-covalent protein–carbohydrate binding sites. We were motivated by the fact that experimental approaches for predicting these sites are expensive. So, computational tools are necessary for identifying these interactions. We explored several sequence-based features as well as structural features. We also leveraged protein language model embeddings. We analyzed different architectures and selected the most suitable deep learning architecture for our finalized prediction model, DeepCPBSite. DeepCPBSite is an ensemble model that combines three separate models with three approaches (random undersampling, weighted oversampling, and class-weighted loss) built on the ResNet+FNN architecture. We made separate datasets from three sources: RCSB, UniProt, and CASP. We also compared the structural features extracted from the structures predicted by AlphaFold and ESMFold in the context of our prediction tasks. We employed three different feature selection techniques and finally did a SHAP (SHapley Additive exPlanations) analysis on the structural features after categorizing the proteins based on their organism information. DeepCPBSite achieved 78.7% balanced accuracy and 59.6% sensitivity on the TS53 set, outperforming the second-best competitor, DeepGlycanSite, by 1.16% and 2.94%, respectively. Additionally, its F1, MCC, and AUPR scores outperformed other state-of-the-art methods, with improvements ranging from 3.77%–47.6%, 3.84%–32.7%, and 8.18%–60.21%, respectively.

|
Scooped by
mhryu@live.com
January 29, 1:01 AM
|
Glycolate, an α-hydroxycarboxylic acid, is widely used in industries such as bioplastics, food, and pharmaceuticals. However, current microbial production methods are limited by the use of plasmids and chemical inducers, hindering their industrial scalability. In this study, a stable and efficient E. coli platform was developed for glycolate production. The glycolate biosynthetic pathway was reconstructed through the identification of a highly efficient glyoxylate reductase (GhrA) from Acetobacter aceti. Carbon flux toward glycolate synthesis was optimized through strategies including enhancing precursor supply, blocking competing pathways, and fine-tuning gene copy numbers. Cofactor engineering was employed by engineering GhrA cofactor preference from NADPH to NADH. Additionally, a non-auxotrophic strain (eliminating exogenous nutrient requirements) for glycolate production was engineered by implementing a growth-stage-dependent molecular switch to dynamically regulate the expression of isocitrate dehydrogenase. Through fermentation optimization, the engineered strain E. coli GA26 achieved a glycolate titer of 81.5 g/L, a yield of 0.49 g/g glucose, and a productivity of 1.9 g/L/h in a 5-L bioreactor, representing the highest reported glycolate titer from glucose to date. These results pave the way for sustainable and cost-effective industrial glycolate production.
|
 Your new post is loading...
Your new post is loading...