Your new post is loading...

|
Scooped by
?
November 7, 5:11 PM
|
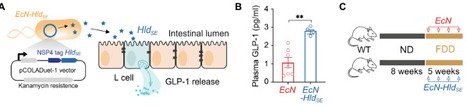
Western diets reduced in fiber promote dysbiosis and exacerbate colitis, with short-chain fatty acids (SCFAs) from fiber fermentation known to regulate glucagon-like peptide 1 (GLP-1) secretion. Whether GLP-1 dysregulation directly links diet-induced dysbiosis to colitis severity, and if this pathway can be therapeutically targeted independently of dietary fiber, remains unclear. Here, we show that dextran sulfate sodium (DSS)–induced colitis severity correlates with compensatory GLP-1 increases, while receptor blockade worsens damage, confirming GLP-1’s protective role during colitis. Fiber deficiency impaired L cell function and GLP-1 release, increasing colitis susceptibility. GLP-1 receptor agonist reversed these effects, restoring barrier integrity and accelerating recovery. We engineered a probiotic, releasing a microbial peptide, that locally elevates GLP-1, which normalized gut parameters in fiber-deprived mice and alleviated colitis via GLP-1–dependent mechanisms, including improved metabolism, antimicrobial defenses, and barrier restoration. Our findings mechanistically connect fiber deficiency to colitis through GLP-1 and demonstrate that probiotic-mediated GLP-1 modulation can bypass dietary fiber requirements to maintain gut homeostasis.

|
Scooped by
?
November 7, 5:05 PM
|
The exploration of post-translational modifications (PTM) within the proteome is pivotal for advancing our understanding of disease and the function of cancer therapeutics. However, identifying genuine sites of PTMs introduced or removed by an enzyme of interest amid numerous candidates is challenging. We present a machine learning (ML)-driven search method, which combines ML with enzyme-mediated modification of complex peptide arrays to predict unexplored PTM sites for an enzyme of interest. Experimental validation confirmed that this approach correctly predicted 37-43% of proposed PTM sites, unveiling candidate sites of the methyltransferase SET8 and the deacetylases SIRT1-7. Our approach marks an important performance increase over traditional in vitro methods across separate enzyme classes. Mass spectrometry analysis confirmed the dynamic methylation status of several predicted SET8 substrates, and the deacetylation of 64 unique sites identified for SIRT2. This method has also revealed changes in SET8-regulated substrate network among breast cancer missense mutations, collectively revealing insight into differential enzyme function in disease. By disentangling the substrate features that dictate PTM-inducing enzyme specificity, this approach demonstrates potential in uncovering enzyme-substrate networks within PTM pathways. Understanding post-translational modifications (PTMs) is crucial for advancing disease research and cancer therapeutics, yet identifying specific enzyme-induced PTM sites remains challenging. Here, the authors introduce a machine learning-driven method combined with high-throughput peptide array synthesis, significantly enhancing PTM site prediction accuracy and revealing insights into enzyme-substrate networks, with implications for cancer research.

|
Scooped by
?
November 7, 4:57 PM
|
Understanding microbial interactions is fundamental for exploring population dynamics, particularly in microbial communities where interactions affect stability and host health. Generalized Lotka-Volterra (gLV) models have been widely used to investigate system dynamics but depend on absolute abundance data, which are often unavailable in microbiome studies. To address this limitation, we introduce an iterative Lotka-Volterra (iLV) model, a novel framework tailored for compositional data that leverages relative abundances and iterative refinements for parameter estimation. The iLV model features two key innovations: an adaptation of the gLV framework to compositional constraints and an iterative optimization strategy combining linear approximations with nonlinear refinements to enhance parameter estimation accuracy. Using simulations and real-world datasets, we demonstrate that iLV surpasses existing methodologies, such as the compositional LV (cLV) and the generalized LV (gLV) model, in recovering interaction coefficients and predicting species trajectories under varying noise levels and temporal resolutions. Applications to the lynx-hare predator-prey, Stylonychia pustula-P. caudatum mixed culture, and cheese microbial systems revealed consistency between predicted and observed relative abundances showcasing its accuracy and robustness. In summary, the iLV model bridges theoretical gLV models and practical compositional data analysis, offering a robust framework to infer microbial interactions and predict community dynamics using relative abundance data, with significant potential for advancing microbial research.

|
Scooped by
?
November 7, 4:46 PM
|
Untargeted tandem mass spectrometry (MS/MS)-based metabolomics enables broad characterization of small molecules in complex samples, yet the majority of spectra in a typical experiment remain unannotated, limiting biological interpretation. Reference data-driven (RDD) metabolomics addresses this gap by contextualizing spectra through comparison to curated, metadata-annotated reference datasets, allowing inference of spectrum origins without requiring exact structural identification. Here, we present an open-source RDD metabolomics platform comprising a user-friendly web application and a Python software package that perform RDD analyses directly from molecular networking outputs generated by GNPS. The tools support visualization and statistical analysis of RDD results, including interactive bar plots, heat maps, principal component analysis, and Sankey diagrams. We illustrate the approach using a hierarchical reference dataset of 3,500 food items to derive dietary patterns from stool metabolomics data of omnivore and vegan participants. The analysis reveals clear dietary group separation, demonstrating how RDD metabolomics can extract biologically meaningful patterns from otherwise unannotated spectra. Thus, the RDD metabolomics platform removes technical barriers for the metabolomics community to adopting reference data-driven analysis, with the functionality freely available at https://github.com/bittremieuxlab/gnps-rdd and https://gnps-rdd.streamlit.app/.

|
Scooped by
?
November 7, 4:34 PM
|
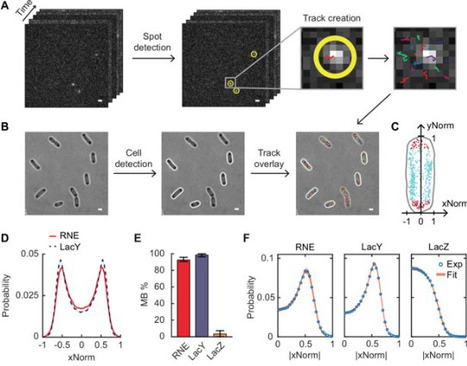
In Escherichia coli, RNase E, a central enzyme in RNA processing and mRNA degradation, contains a catalytic N-terminal domain, a membrane-targeting sequence (MTS), and a C-terminal domain (CTD). We investigated how MTS and CTD influence RNase E localization, diffusion, and function. Super-resolution microscopy revealed that ~93% of RNase E localizes to the inner membrane and exhibits slow diffusion similar to polysomes. Comparing the native amphipathic MTS with a transmembrane motif showed that the MTS confers slower diffusion and stronger membrane binding. The CTD further slows diffusion by increasing mass but unexpectedly weakens membrane association. RNase E mutants with partial cytoplasmic localization displayed enhanced co-transcriptional degradation of lacZ mRNA. These findings indicate that variations in the MTS and the presence of the CTD shape the spatiotemporal organization of RNA processing in bacterial cells, providing mechanistic insight into how RNase E domain architecture influences its cellular function.

|
Scooped by
?
November 7, 4:21 PM
|
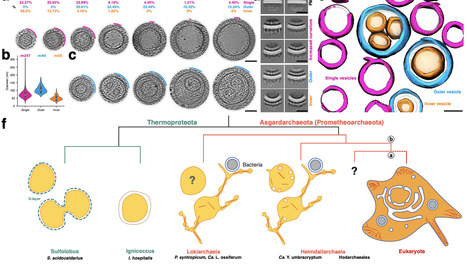
The emergence of eukaryotes from a merger between an archaeon and a bacterial cell ~two billion years ago involved a profound change in cellular organization. While the order in which different features of the eukaryotic cell arose remains a matter of controversy, close archaeal relatives of eukaryotes have recently been identified that possess homologues of eukaryotic trafficking machinery and a complex cell architecture. The members of this phylum, the Asgard archaea (syn. Prometheoarchaeota) described so far, however, lack internal membrane-bound compartments, and therefore have shed little light on origins of the hallmark eukaryotic endomembrane system. Here we report the cell biological analysis of a member of the Heimdallarchaeia, Candidatus "Yibarchaeum umbracryptum" in enriched mixed microbial communities. Possessing a small genome encoding few homologues of eukaryotic membrane remodelling machinery, Ca. Y. umbracryptum cells in late-stage cultures resemble previously described Asgard archaea with extensive cellular protrusions. Surprisingly, during early stages of culture growth Ca. Y. umbracryptum cells have fewer protrusions but possess numerous intracellular vesicles, most of which have a luminal surface that morphologically resembles the outer coat of the plasma membrane. These data alter our view of eukaryogenesis by identifying a close archaeal relative of eukaryotes with a regulated endomembrane system.

|
Scooped by
?
November 7, 3:09 PM
|
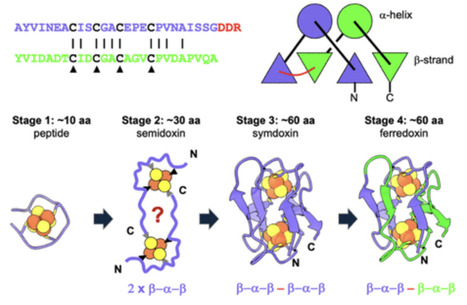
Electron transfer coupled to redox chemistry is at the heart of metabolism. The proteins responsible for moving electrons (protein electron carriers) must have emerged at the origin of life. The small iron–sulfur-binding bacterial ferredoxins were likely among these first proteins. Embedded within the ferredoxin sequence and structure is a symmetry that points to an ancient gene duplication event. Little is understood about the nature of ferredoxins prior to this duplication event or what environmental factors may have driven the selection for more complex forms. The deep-time molecular history of ferredoxins goes back billions of years and cannot be reconstructed by phylogenetic analyses based on amino acid sequences. Here, we use structure-guided protein design to model a fossil half-ferredoxin stage in the evolution of this fold, the semidoxins, and their symmetric full-length counterparts, the symdoxins. Semidoxin designs homodimerize, exhibiting structural, thermodynamic, and electrochemical behaviors in most cases identical to cognate symdoxins. However, the semi- and symdoxin fossil stages behave differently when incorporated into an in vivo electron transfer complementation assay. Both can support bacterial growth dependent on protein expression. Growth rates of bacteria expressing the semidoxins are much more sensitive to oxygen than those of bacteria expressing symdoxins. Motivated by the in vivo functionality of designed semidoxins, we identified putative naturally occurring semidoxins in extant anaerobic microorganisms. This is consistent with the observed in vivo oxygen sensitivity of the semidoxin designs. One natural semidoxin is shown to be folded and redox active. However, it exists as a mixture of monomers and dimers, suggesting a potential connection between semidoxins and even simpler single iron–sulfur cluster-binding peptides.

|
Scooped by
?
November 7, 2:21 PM
|
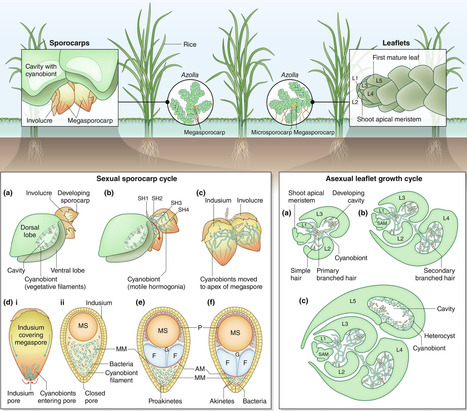
Heritable symbioses exist across eukaryotes with different degrees of intimacy. In most cases, the symbionts are obligate and require inheritance for their survival. On the host side, symbiont retention can facilitate fitness benefits. Only rarely are these symbioses interwoven to the point that host survival relies on the symbiont. In land plants, the symbiosis of the water fern Azolla with its symbiotic cyanobacterium shows such a degree of high co-dependence. The symbiosis originated in the last common ancestor of Azolla and exists continuously for at least 60 million years with no evolutionarily stable, secondary loss of the symbiont reported. This is a feat achieved by interactions on an organellar-like level or those considered recent organelle acquisitions. Yet, Azolla's symbiont is extracellular. How can loss of autonomy concomitant with full co-dependence be accommodated in this extracellular symbiosis? Here, we synthesize what we know from the Azolla symbiosis on the consequences of evolutionary co-dependence and stable symbiont retention. We discuss the need for symbiotic integration into environmental responsiveness if host survival depends on symbiont well-being. Cross-organismal integration of environmental stress responses may be one of the key steps that favor this evolutionarily stable permanent integration.

|
Scooped by
?
November 7, 1:56 PM
|
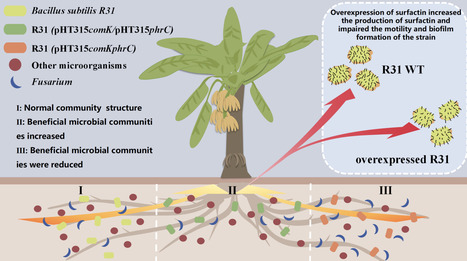
Surfactin, a lipopeptide antibiotic and quorum-sensing (QS) mediator from Bacillus subtilis, has dual functions in microbial ecology and plant disease suppression. This study engineered B. subtilis R31 to overproduce comK and phrC, key regulators of surfactin biosynthesis, increasing surfactin yield by 45% compared to the WT strain. While elevated surfactin enhanced antimicrobial potential, comK-mediated overproduction impaired biofilm formation and swarming motility, but rhizosphere colonization was mostly unaffected. 16S rRNA sequencing of banana rhizospheres showed that surfactin selectively shaped the microbial community by enriching beneficial Bacillus species. Mechanistic studies confirmed surfactin's dual role as an antimicrobial and an intercellular signalling molecule for coordinated development in Bacillus populations. These results reveal the molecular mechanisms of R31-mediated suppression of banana Fusarium wilt and offer a strategy for engineering synthetic microbial consortia by manipulating metabolic signalling pathways.

|
Scooped by
?
November 7, 11:48 AM
|
Microorganisms commonly live in diverse communities where changes in composition can be critical for health, industry and the environment. Yet, what enables one strain to competitively replace another in these complex conditions remains poorly understood. Here we develop a mathematical model to determine general principles of strain displacement. Our modelling reveals that weak resource competition enables successful invasion while strong interference competition, for example, via antimicrobial production, enables successful displacement. We verify these predictions using in vitro assays with genetically engineered Escherichia coli. We then apply our principles to displace multidrug-resistant clinical isolates using strains that are equipped with a potent bacteriocin. Finally, we perform experiments with diverse human gut symbionts, which reveal that displacement relies on low resource competition not only between competing strains but also with the broader community, that is, limited nutrient blocking. These general rules for ecological success in microbial communities could be applied for targeted displacement of bacteria. Mathematical modelling and experimental tests reveal principles that govern displacement of a resident strain by an invader in microbial communities.

|
Scooped by
?
November 7, 1:30 AM
|
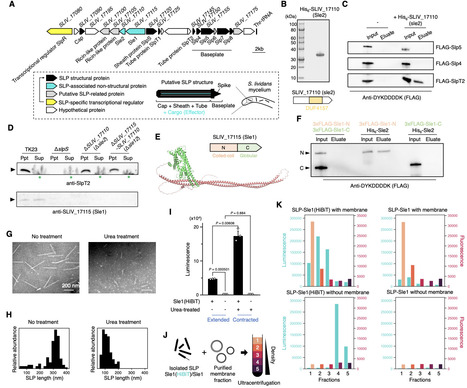
Contractile injection systems (CISs) are derivatives of phage tails and widely distributed in prokaryotes. CISs load cognate effectors and eject them through contractile actions resembling those of phage tails. Ejected effectors play central roles in CIS functionality by acting on target cells and mediating various biological processes. Here, we report a novel group of CIS effectors related to phage tapemeasure protein, the transmembrane component of the phage infection machinery. This group is broadly distributed within the class actinobacteria, one of the bacterial classes in which CIS gene clusters are highly conserved, and is represented by Sle1, a cognate effector of the intracellularly localised Streptomyces lividans phage tail-like nanoparticle (SLP). This effector is associated with Sle2, which contains a CIS effector core domain and interacts with the SLP core component. Sle1 is packaged inside SLP and is translocated to lipid membranes along with SLPs. The functional domain of Sle1, probably through interactions with ribosome-containing subcellular fractions, upregulates the membrane-associated proteome in S. lividans and E. coli. This effect modifies the physiological properties of S. lividans, ultimately enhancing its adaptation to microbial competition. In addition, we revealed that Sle1-type effectors conserved among actinobacterial species are structurally and functionally diverse in their functional domains. One of them from Micromonospora eburnea constitutes a novel toxin-antitoxin system and introducing its functional domain into Sle1 reprogrammes the phenotypic responsiveness of S. lividans to neighboring bacteria. Our findings illustrate that phage elements can be incorporated into CISs as reconfigurable platforms for bacterial adaptation to various environmental conditions.

|
Scooped by
?
November 7, 12:10 AM
|
Miniature inverted-repeat transposable elements (MITEs) are nonautonomous mobile genetic elements (MGEs) that can be mobilized by transposases provided by the relevant autonomous MGEs. Composite transposon-like structures bounded by two MITE copies, designated MITE-COMPs, were previously shown to mobilize the intervening sequences. However, their association with antibiotic-resistance genes (ARGs) has not yet been systematically studied, and only a few MITE-COMP structures with ARGs have been reported to date. This study thus aimed to identify new MITE-COMP members containing ARGs in the genomic sequences of the clinically important bacterial family Enterobacteriaceae in public databases. MITE-COMPs were first searched for in the PLSDB database, focusing on small plasmids, using a self-against-self blastn approach. Then, newly and previously identified MITEs were searched for in the NCBI core nucleotide database to identify MITE-COMPs located on other replicons. Bioinformatics analyses identified multiple previously unreported MITE-COMP structures containing ARGs, which are bounded by directly or inversely oriented MITEs, including IS101, MITESen1, and a novel 244-bp MITE termed MITE244. All these MITEs seem to have originated from transposons of the Tn3 family. MITE244 contains a putative cointegrate resolution site related to Tn21, and MITE244 copies in MITE-COMPs were predominantly found in inverse orientation. The ARGs embedded in newly identified MITE-COMPs include blaKPC-2, qnrS1, tet(A), qnrD1, and floR. Notably, the blaKPC-2 carbapenemase gene was found in MITE-COMPs bounded by MITE244 and another MITE-COMP bounded by IS101. These findings highlight a potential role for MITEs in the spread of diverse ARGs in Enterobacteriaceae.

|
Scooped by
?
November 7, 12:00 AM
|
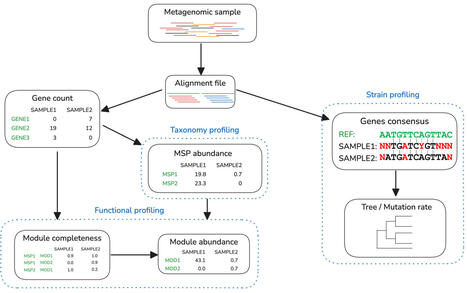
The characterization of complex microbial communities is a critical challenge in microbiome research, as it is essential for understanding the intricate relationships between microorganisms and their environments. Metagenomic profiling has advanced into a multifaceted approach, combining taxonomic, functional, and strain-level profiling (TFSP) of microbial communities. Here, we present Meteor2, a tool that leverages compact, environment-specific microbial gene catalogues to deliver comprehensive TFSP insights from metagenomic samples. Meteor2 currently supports 10 ecosystems, gathering 63,494,365 microbial genes clustered into 11,653 metagenomic species pangenomes (MSPs). These genes are extensively annotated for KEGG orthology, carbohydrate-active enzymes (CAZymes) and antibiotic-resistant genes (ARGs). In benchmark tests, Meteor2 demonstrated strong performance in TFSP, particularly excelling in detecting low-abundance species. When applied to shallow-sequenced datasets, Meteor2 improved species detection sensitivity by at least 45% for both human and mouse gut microbiota simulations compared to MetaPhlAn4 or sylph. For functional profiling, Meteor2 improved abundance estimation accuracy by at least 35% compared to HUMAnN3 (based on Bray–Curtis dissimilarity). Additionally, Meteor2 tracked more strain pairs than StrainPhlAn, capturing an additional 9.8% on the human dataset and 19.4% on the mouse dataset. Furthermore, in its fast configuration, Meteor2 emerges as one of the fastest available tools for profiling, requiring only 2.3 min for taxonomic analysis and 10 min for strain-level analysis against the human microbial gene catalogue when processing 10 M paired reads — operating within a modest 5 GB RAM footprint. We further validated Meteor2 using a published faecal microbiota transplantation (FMT) dataset, demonstrating its ability to deliver an extensive and actionable metagenomic analysis. The unified database design also simplifies the integration of TFSP outputs, making it straightforward for researchers to interpret and compare results. These results highlight Meteor2 as a robust and versatile tool for advancing microbiome research and applications. As an open-source, easy-to-install, and accurate analysis platform, Meteor2 is highly accessible to researchers, facilitating the exploration of complex microbial ecosystems.
|

|
Scooped by
?
November 7, 5:08 PM
|
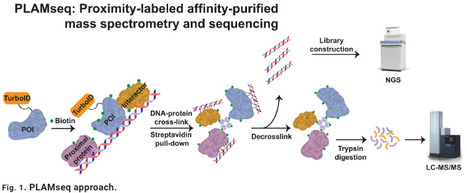
Chromatin immunoprecipitation and coimmunoprecipitation assays are common approaches to characterize the genomic localization and protein interactors, respectively, for a protein of interest. However, these approaches require the use of specific antibodies, which often face sensitivity and specificity issues. On the basis of TurboID, we developed proximity-labeled affinity-purified mass spectrometry and sequencing (PLAMseq), which enables, in the same workflow, identification of the genomic loci and the interacting proteome of a protein of interest. Moreover, PLAMseq can also be applied to specifically map protein interactions and ubiquitin(-like)–modified proteins. We validated PLAMseq with two well-characterized proteins, RNA polymerase II, and CTCF, with excellent robustness and reproducibility. Next, we applied PLAMseq to characterize histone H1 SUMOylation, in which study has remained elusive due to the lack of specific reagents, and found that SETDB1 binds to SUMOylated histones H1.2 and H1.4 that also colocalize with H3K9me3 at repetitive regions of the genome.

|
Scooped by
?
November 7, 5:01 PM
|
Protein solubility is a critical physicochemical property influencing protein stability, therapeutic efficacy, and overall developability in drug discovery. However, traditional experimental methods for assessing solubility are often resource-intensive and time-consuming. To address these limitations, computational approaches leveraging artificial intelligence have emerged, yet current models generally treat qualitative classification and quantitative regression as separate tasks and rely predominantly on sequence-based information, neglecting crucial structural and surface characteristics. Here, we introduce Pro4S, a novel multimodal predictive model that integrates protein language models, structural data, and surface descriptors using advanced contrastive learning techniques. Our unified framework achieves significant improvements in prediction accuracy, robustness, and generalizability for both qualitative and quantitative solubility assessments. Benchmark comparisons demonstrate that Pro4S consistently outperforms existing state-of-the-art predictors across diverse datasets. Furthermore, by applying Pro4S to the emerging area of de novo protein design, we validated a strong correlation between predicted solubility and experimental expression levels, reducing the proportion of non-expressed proteins by 52.7% while retaining 96.7% of highly expressed proteins. This highlights Pro4S's potential to serve as a reliable upfront screening tool for increasing expression success rates and accelerating rational protein engineering.

|
Scooped by
?
November 7, 4:53 PM
|
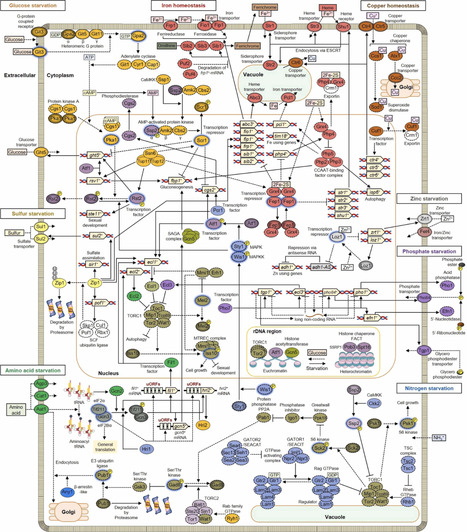
In nature, nutrient-poor environments are more common than exposure to nutrient-rich environments, and living organisms have developed countermeasures to survive nutrient starvation. Increasing research has revealed beneficial aspects of starvation for an individual’s life, including lifespan extension. The fission yeast Schizosaccharomyces pombe is a model unicellular eukaryotic organism and has greatly contributed to the understanding of various cellular processes, including the cell cycle, cell morphology, sexual development, cell lifespan, and nutritional responses. Traditionally, research on starvation in fission yeast has focused on glucose starvation and nitrogen starvation. Recently, studies on cellular responses to the starvation of various nutrients, such as phosphorus, sulfur, iron, zinc, copper, and amino acids have been reported, revealing similarities and differences among the various types of nutrient starvation. In fission yeast, Ecl proteins, which are conserved among fungi, can sense the starvation of multiple nutrients. These proteins also repress the target of rapamycin complex 1 (TORC1), which is conserved across eukaryotes. They channel a variety of starvation signals into common cellular responses, such as growth arrest, sexual differentiation, autophagy, and lifespan extension. This review summarizes and discusses the signaling mechanisms involved in the initial cellular responses of fission yeast to the starvation of various nutrients.

|
Scooped by
?
November 7, 4:39 PM
|
The development of hydrogenases capable of operating under oxygenic photosynthetic conditions remains a key challenge for sustainable biohydrogen production. In this study, we developed a series of chimeric NAD+-reducing [NiFe]-hydrogenases (SH) combining structural elements from the O2-tolerant SH of Cupriavidus necator (CnSH) and the ferredoxin-interacting SH of Synechocystis sp. PCC6803 (SynSH). By engineering chimeric HoxU and HoxF subunits, we developed constructs─MixSH, Ch-HoxEFSyn+UCn, and Ch-HoxUswapCTD─that successfully couple the CnHoxYH hydrogenase module to the SynHoxEFU reductase module while retaining O2 tolerance and enhancing interaction with reduced ferredoxin. The lithoautotrophic growth of C. necator confirmed the tolerance of these variants to O2, while activity assays in Synechocystis demonstrated partial hydrogenase function, including H2 consumption and fermentative H2 production. Notably, Ch-HoxEFSyn+UCn retained ferredoxin interaction despite lacking the [4Fe4S]U4 cluster, showing [2Fe2S]F2 in HoxF as a functional ferredoxin-binding site. Moreover, we achieved the artificial integration of a [2Fe2S] cluster into CnHoxF and identified the CnHoxF N-terminal domain as structurally and functionally analogous to SynHoxE. Although electron transfer efficiency and activity in Synechocystis remained limited, this work validates the modular engineering of [NiFe]-hydrogenases, uniting O2-tolerance with ferredoxin interaction and offering a foundational step toward photosynthesis-coupled H2 production.

|
Scooped by
?
November 7, 4:30 PM
|
Multiple global change drivers have caused a large carbon (C) debt in our soils. To remedy this debt, understanding the role of microorganisms in soil C cycling is crucial to tackle the C soil loss. Microbial carbon use efficiency (CUE) is a parameter that captures the formation of microbially-derived soil organic matter (SOM). While it is known that biotic and abiotic drivers influence CUE, it remains unclear whether bacteria, fungi and their interactions influence the formation of microbially-derived SOC and its persistence in soils. Here, we combined the inoculation of distinct communities (a biotic factor) grown at different moisture levels (an abiotic factor) to manipulate the formation of microbial necromass in model soil. In a follow-up experiment, we then evaluated the persistence of this previously formed microbially-derived C to decomposition. While we show that necromass formation reflects the microbial community composition, the SOC formed within the most complex community of bacteria and fungi seems to be more resistant to decomposition compared to the SOC formed within the simpler communities (bacteria and fungi simple community, bacteria only and fungi only communities). Moreover, fungal necromass proved to be more thermally-stable than bacterial necromass, if this necromass is formed with both bacteria and fungi present. Our findings reveal that although abiotic factors can influence microbial physiology, the biological origin of microbially-derived C and the co-occurrence of fungal and bacterial growth were the stronger drivers explaining SOM persistence in these soils, suggesting the importance of microbial succession in SOC stabilization.

|
Scooped by
?
November 7, 3:13 PM
|
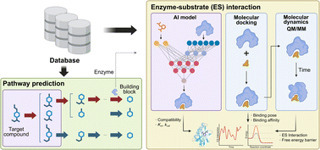
Natural products are a major source of bioactive compounds, yet elucidating their biosynthetic pathways remains a major challenge due to complex genotype–phenotype relationships. Recent advances in computational approaches, particularly artificial intelligence (AI) and mechanistic modeling, are transforming this field. This highlight examines key databases that underpin computational studies, AI-driven methods for predicting biosynthetic pathways and enzyme–substrate interactions, and mechanistic simulations that provide energetic and structural insights. We also discuss current challenges and future opportunities for integrating these strategies to accelerate discovery, engineering, and application of natural products in drug discovery, biotechnology, and synthetic biology.

|
Scooped by
?
November 7, 2:22 PM
|
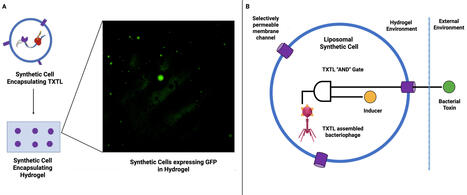
A synthetic cell is a membrane-bound vesicle that encapsulates cell-free transcription/translation (TXTL) systems. It represents a transformative platform for advancing bacteriophage therapy. Building on experimental work that demonstrates (i) modular genome assembly, (ii) high-yield phage TXTL systems, and (iii) smart hydrogel encapsulation, we explore how synthetic cells can address major limitations in phage therapy. The promising advances include point-of-care phage manufacturing, logic-responsive antimicrobial biomaterials, and new chassis to dissect the dynamics of phage-host interactions. We also propose a roadmap for the deployment of synthetic cells as programmable and evolvable tools in the context of laboratory research and translational clinical adoption. droplet

|
Scooped by
?
November 7, 2:01 PM
|
This work describes the development of a CRISPR interference (CRISPRi) system for targeted gene repression in bifidobacteria. We first validated the CRISPRi-based approach using Bifidobacterium breve strains engineered to express nuclease-dead orthologs of Cas9 and demonstrated that the CRISPR-Cas system from Streptococcus thermophilus is efficient at targeting both reporter and endogenous genes through the use of single guide RNAs corresponding to the gene of interest. We also developed a one-plasmid system for targeted gene repression in bifidobacteria and demonstrated its utility by targeting genes involved in nucleotide metabolism and carbohydrate metabolism in several species of bifidobacteria. Efficient gene repression was achieved across all tested bifidobacterial species without the requirement for extensive optimization of transformation parameters or sequence optimization to avoid restriction modification systems thus removing the key barriers to genetic manipulation in this genus. This CRISPRi system provides a novel approach to functional genomics in bifidobacteria which facilitates future mechanistic studies in these commercially important microbes.

|
Scooped by
?
November 7, 12:52 PM
|
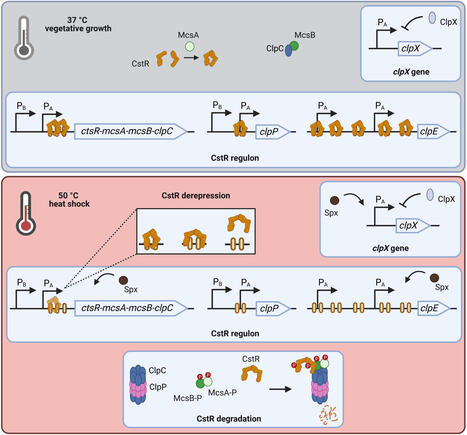
A sudden increase in temperature triggers Bacillus subtilis to activate expression of stress-specific heat shock proteins of the CtsR (class three stress gene repressor) regulon to withstand the adverse conditions. Key members of this regulon, such as ATPases, proteolytic subunits and their adaptors, which can assemble to the functional Clp protease system, perform crucial roles in maintaining cellular proteostasis, while their transcription is repressed by CtsR during vegetative growth. Upon heat shock, a conformational change in a thermo-sensing glycine-rich loop causes CtsR to detach from its DNA operators, enabling the transcriptional activation of the regulon. Novel data from a clpX-deficient strain demonstrated that in addition, the presence of the ATPase ClpX is essential for the CtsR dissociation from its DNA binding site. To further elucidate this role of ClpX, we constructed a conditional clpX strain, in which clpX induction is decoupled from its native transcriptional control. This conditional expression system mimicked a clpX-deficient phenotype under non-inducing conditions and restored the wild-type phenotype upon induction. Our results indicate that the full induction of the CtsR regulon, particularly clpE, requires both heat and the presence of ClpX, thereby extending the current model for the transcriptional activation of genes repressed by CtsR.

|
Scooped by
?
November 7, 9:38 AM
|
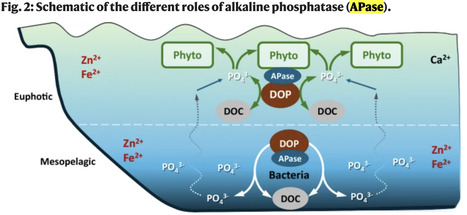
Phosphorus availability is vital to ocean ecosystem functioning. Analyses of global seawater samples combined with laboratory experiments reveal the Alteromonas bacteria as a surprising regulator of the marine phosphorus cycling.

|
Scooped by
?
November 7, 12:20 AM
|
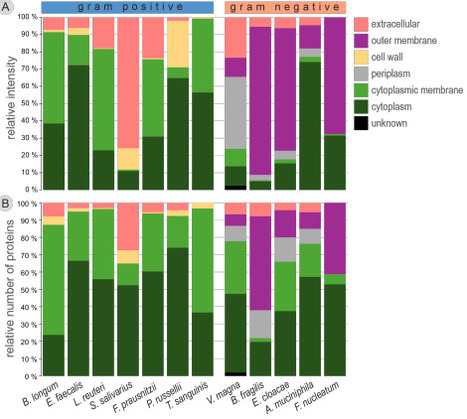
Bacterial extracellular vesicles (bEVs) produced by intestinal commensal bacteria mediate host-microbe interactions, but their proteomes have been explored in only a small number of species. We characterized bEVs from in vitro cultures of 12 species of Actinomycetota, Bacillota (Firmicutes), Bacteroidota, Fusobacteria, Pseudomonadota, and Verrucomicrobia. This is the first report of bEVs from Veillonella magna, an exceptional gram-negative Bacillota, and the gram-positives Peptostreptococcus russellii and Turicibacter sanguinis. The morphology and protein subcellular localization patterns of bEVs reflected the envelope structures of their parent bacteria. Notably, V. magna may be able to produce both outer membrane and cytoplasmic membrane vesicles. The proteome compositions were dictated by phylogeny, suggesting largely non-selective packaging of proteins. Annotation of protein functions indicated roles in nutrient metabolism and transport, and in host immune system modulation. Peptidoglycan modifying enzymes and the abundant bacteriophage proteins in Enterobacter cloacae and Limosilactobacillus reuteri bEVs may be involved in vesicle biogenesis. The functions of many abundant bEV proteins are unknown. The most abundant bEV proteins were largely species-specific, but we identified several conserved proteins that may be used as markers to distinguish commensal bEVs from host EVs. Comparison to previous in vitro and fecal metaproteomics data indicates that bEV proteome compositions are reproducible.

|
Scooped by
?
November 7, 12:01 AM
|
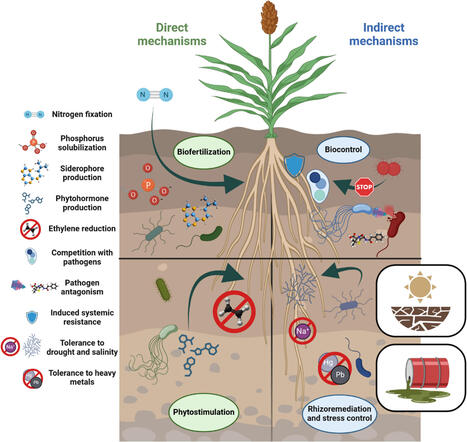
This review highlights the benefits of mutualistic plant–microbe interactions in enhancing the resilience of emergent crops and underlies the potential of these crops as valuable resources for exploring novel plant growth-promoting bacteria (PGPB). Emergent crops such as quinoa, amaranth, millet, lupins, hemp and desert truffles exhibit physiological and ecological traits that make them suitable for stress-prone environments. PGPR offer sustainable solutions to mitigate abiotic stress by improving nutrient availability, modulating phytohormone levels, enhancing root development and inducing systemic resistance. While their benefits have been extensively documented in model crops under controlled conditions, their application in emergent crops remains underexplored. This review examines current knowledge on the individual and combined roles of these crops and microbes, highlighting specific examples where PGPB have improved plant performance, yield and stress tolerance. Microbial inoculants show potential not only to boost productivity but also to reduce agrochemical inputs, contributing to sustainability. The review also discusses the need for tailored microbial formulations and effective field application strategies to bridge the gap between experimental research and real-world agricultural practices. Identifying key knowledge gaps, it emphasizes the importance of further research on strain specificity, crop-microbe interactions and multi-strain consortia for scalable and climate-smart agriculture. Ultimately, harnessing the synergisms between PGPB with emergent crops could convert marginal lands into productive, climate-smart farms, increasing food security in the face of environmental challenges posed by global climate change.
|
 Your new post is loading...
Your new post is loading...