Your new post is loading...

|
Scooped by
?
June 26, 11:49 PM
|
Genomic methylation in bacteria plays a crucial role in gene regulation, chromosome replication, pathogenicity, and defence against phages. While single-molecule real-time (SMRT) sequencing technologies have advanced the detection of epigenetically modified bases, the statistical analysis of their distribution and the possible roles they play in bacterial cells remains challenging. To address this gap, we developed SeqWord Motif Mapper (SWMM), a computational tool designed for the statistical analysis and visualization of bacterial methylation patterns. SWMM utilizes PacBio sequencing data to identify sequence coverage, methylation motif distribution, and putative functional associations. Implemented in Python 3.9, the tool is platform-independent and requires minimal dependencies, making it accessible to a wide range of users. The SWMM command-line interface and a web-based version of the program facilitate the exploration of epigenetic modifications across bacterial genomes. Through case studies on different bacterial and archaeal taxa, we demonstrated that genome methylation in microorganisms extends beyond canonical sites and possibly influences gene expression, adaptation, and genome architecture. The tool enables detailed statistical evaluation of methylation motif distribution and provides insights into the potential regulatory roles of epigenetic modifications in bacterial genomes. SWMM is freely available at https://begp.bi.up.ac.za, with source code hosted on GitHub at https://github.com/chrilef/BactEpiGenPro.

|
Scooped by
?
June 26, 11:39 PM
|
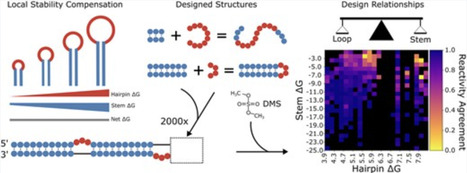
RNA molecules adopt complex structures that perform essential biological functions across all forms of life, making them promising candidates for therapeutic applications. However, our ability to design new RNA structures remains limited by an incomplete understanding of their folding principles. While global metrics such as the minimum free energy are widely used, they are at odds with naturally occurring structures and incompatible with established design rules. Here, we introduce local stability compensation (LSC), a principle that RNA folding is governed by the local balance between destabilizing loops and their stabilizing adjacent stems, challenging the focus on global energetic optimization. Analysis of over 100 000 RNA structures revealed that LSC signatures are particularly pronounced in bulges and their adjacent stems, with distinct patterns across different RNA families that align with their biological functions. To validate LSC experimentally, we systematically analyzed thousands of RNA variants using DMS chemical mapping. Our results demonstrate that stem folding, as measured by reactivity, correlates with LSC (R² = 0.458 for hairpin loops) and that instabilities show no significant effect on folding for distal stems. These findings demonstrate that LSC can be a guiding principle for understanding RNA function and for the rational design of custom RNAs.

|
Scooped by
?
June 26, 11:22 PM
|
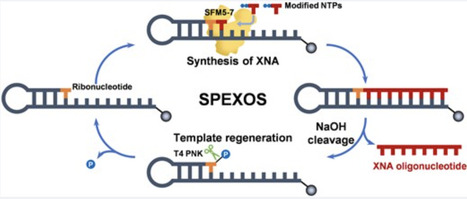
Xenobiotic nucleic acids (XNAs) significantly expand the range of genetic polymers and serve as promising alternatives to DNA and RNA for numerous biological applications. However, the extensive exploration and application of XNAs are limited by low sustainability and yields in solid-phase oligonucleotide synthesis, as well as by the unavailability of efficient XNA polymerases for polymerase-mediated XNA production. To address the limitations in XNA production, we developed a solid-phase enzymatic XNA oligonucleotide synthesis platform using a laboratory-evolved XNA polymerase, SFM5-7, which exhibits excellent activity for synthesizing DNA, RNA, 2′-fluoroarabinonucleic acid (FANA), and other 2′-modified XNA oligonucleotides. This platform employs ribonucleotide insertion and alkaline cleavage of the oligonucleotide product before and after SFM5-7-mediated XNA synthesis, enabling recycled XNA synthesis through the reuse of a bead-immobilized self-priming hairpin DNA template. The platform’s potential and versatility are demonstrated by the production of FANA, 2′-modified RNAs, chimeric XNAs, 5′-end-labeled FANA, and an active FANAzyme. This platform should facilitate the customized production of functional XNAs with programmable modifications, accelerating their applications in biotechnology and biomedicine.

|
Scooped by
?
June 26, 1:44 AM
|
Protein–protein interactions ppi are no longer considered undruggable because of the conceptual and technical advances that allow inhibitors to be generated using rational design principles and high-throughput screening methods. Here we review the concepts and approaches that have underpinned the progress in this field. We begin by assessing what makes a protein surface more tractable than others with a focus on the recent success in targeting Ras, which has long served as a poster child of a therapeutically important yet undruggable target. We discuss computational approaches to dissect protein surfaces to design macrocycles and miniprotein ligands. Traditional drug discovery has benefitted from leveraging natural products but this benefit has not extended to the design of ligands for protein surfaces because few natural products have been characterized as inhibitors of protein complexes. However, nature does provide a template in the form of binding epitopes of partner proteins. We review design of protein structure mimics that enable rational design of inhibitors through multiple weak contacts. Lastly, we focus on contemporary screening methods that are being merged with constrained peptides to offer unprecedented side chain diversity on conformationally defined scaffolds. We will focus on the concepts underlying advancements in the field rather than the application of these concepts and technologies that have led to inhibitors of specific interactions.

|
Scooped by
?
June 26, 1:38 AM
|
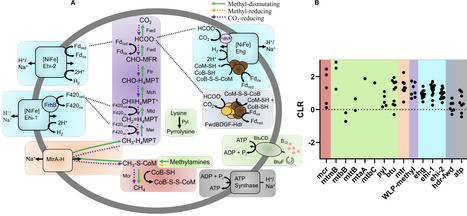
Methane is a potent greenhouse gas that is largely produced through the activity of methanogenic archaea, which contribute to Earth's dynamic climate and biogeochemical cycles. In the past decade, metagenomics revealed that lineages outside of the traditional Euryarchaeota superphylum encode genes for methanogenesis. This was recently confirmed through the cultivation of two classes of methanogenic Thermoproteota. Thus far, all methanogens within the Thermoproteota are predicted or were shown to be methylotrophic. The only exception to this is the Nezhaarchaea, which, based on metagenomics, are predicted to be CO2-reducing methanogens. Here, we demonstrate methanogenic activity in a third class of Thermoproteota, the Methanonezhaarchaeia. We expand the metabolic diversity of this class by cultivating a methylotrophic species, Candidatus Methanonezhaarchaeum fastidiosum YNP3N. We describe novel genes involved in methanogenesis that are not found in other methanogens. We investigate the metabolic diversity of Methanonezhaarchaeia, including metabolic modifications accompanying frequent loss of methanogenesis in this class. This highlights gaps in our understanding of the biochemistry, diversity, and evolution of non-traditional methanogens and their contributions to carbon cycling.

|
Scooped by
?
June 26, 1:34 AM
|
Translational termination is not entirely efficient and competes with elongation, which might result in translational readthrough (TR). TR occurs when a near-cognate tRNA binds to a stop codon, (mis)interpreting it as a sense codon and producing a C-terminal extension of the protein. This process is influenced by the stop codon itself and the surrounding nucleotide sequence, known as the stop codon context (SCC). To investigate the role of these cis-acting elements beyond the high-TR motif UGA CUA G, this study examines specific positions within the SCC, both upstream and downstream of the motif, that contribute to variations in basal and aminoglycoside-induced TR. In particular, we identified a surprisingly large influence of the upstream nucleotide positions -9 and -8 (relative to the stop codon) and positions +11 and +12 on readthrough levels, revealing a complex interplay between nucleotides in the expanded SCC with effects turning out to be non-linear and, furthermore, not transferable to evolutionarily non-adapted SCCs. These findings support our understanding of translational termination and may benefit the development of pharmacological therapy for diseases caused by premature stop codon mutations.

|
Scooped by
?
June 26, 1:28 AM
|
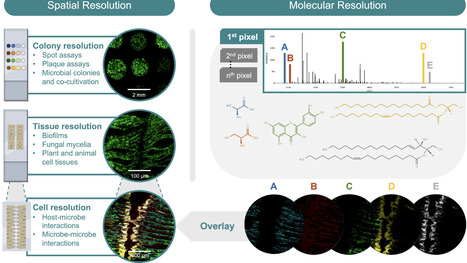
Spatial metabolomics is transforming our understanding of microbial systems by uncovering the localized molecular dynamics within complex microbial communities. By using advanced mass spectrometry imaging techniques, researchers can now visualize metabolites at the micron scale and show how microbes interact, form biofilms, and influence their environments at unprecedented levels of detail. These approaches not only provide molecular insights they also highlight the need of integrating activity measurements and temporal analyses to capture the dynamic nature of microbial assemblages. Incorporating these dimensions in future will enable a deeper understanding of microbial interactions over time, bridging the gap between static snapshots and the evolving processes that drive microbial ecosystems. This commentary explores important aspects in microbial spatial metabolomics, underscores the need for activity and time-resolved studies, and advocates for a deeper exploration of microbial systems to advance chemical ecological concepts, host–microbe interactions, and metabolic mechanisms in microbial communities.

|
Scooped by
?
June 26, 1:20 AM
|
Legumes are not only major cash crops but also contribute valuable nitrogen to cropping systems due to their ability to form a symbiotic relationship with nitrogen-fixing rhizobia in specialized root organs called nodules. To balance the cost of carbon provision to the rhizobia, nodulation is finely regulated in legumes across various spatiotemporal levels, including host–microbe signalling within the rhizosphere, infection of the legume host, and nodule initiation, function, and senescence. Since symbiotic nitrogen fixation (SNF) evolved in natural ecosystems which lack resemblance to modern agricultural systems, opportunities present themselves to genetically improve SNF. Based on recent findings and the opportunities arising with new breeding technologies, we review here the many opportunities to optimise SNF and highlight the key challenges associated with these approaches.

|
Scooped by
?
June 26, 1:13 AM
|
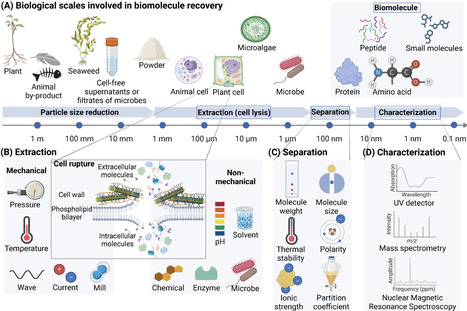
The growing demand for sustainable solutions in agriculture, which are critical for crop productivity and food quality in the face of climate change and the need to reduce agrochemical usage, has brought biostimulants into the spotlight as valuable tools for regenerative agriculture. With their diverse biological activities, biostimulants can contribute to crop growth, nutrient use efficiency, and abiotic stress resilience, as well as to the restoration of soil health. Biomolecules include humic substances, protein lysates, phenolics, and carbohydrates have undergone thorough investigation because of their demonstrated biostimulant activities. Here, we review the process of the discovery and development of extract-based biostimulants, and propose a practical step-by-step pipeline that starts with initial identification of biomolecules, followed by extraction and isolation, determination of bioactivity, identification of active compound(s), elucidation of mechanisms, formulation, and assessment of effectiveness. The different steps generate a roadmap that aims to expedite the transfer of interdisciplinary knowledge from laboratory-scale studies to pilot-scale production in practical scenarios that are aligned with the prevailing regulatory frameworks.

|
Scooped by
?
June 26, 12:48 AM
|
With the sustainable increase in agricultural productivity, the need for safer, environmentally friendly pesticide alternatives is also growing. Metabolites of microorganisms (bacteria, fungi, actinomycetes) are emerging as potential bioactive compounds for integrated pest and disease management. These compounds comprise amino acids, carbohydrates, lipids, organic acids, phenolics, peptides, alkaloids, polyketides, and volatile organic compounds. The majority of them have insecticidal, fungicidal, and nematicidal activities. In this review, the classifications, biosynthetic pathways, and ecological functions of primary and secondary metabolites produced by microorganisms are discussed, including their mechanisms of action, ranging from competition to systemic acquired resistance in host plants. The article highlights the importance of microbial genera (viz., Bacillus sp., Pseudomonas sp., Trichoderma sp., Streptomyces sp., etc.) in making chemicals and biopesticides for crop defense. We present the possible applications of microbial biosynthesis strategies and synthetic biology tools in bioprocess development, covering recent innovations in formulation, delivery, and pathway engineering to enhance metabolite production. This review emphasizes the significance of microbial metabolites in improving the plant immunity, yield performance, reduction in pesticide application, and the sustainability of an ecological, sustainable, and resilient agricultural system.

|
Scooped by
?
June 26, 12:45 AM
|
Most bacteriophages lyse their host cell to release progeny virions. Double-stranded DNA phages typically promote host lysis using a holin-endolysin system. Holins form pores in the cytoplasmic membrane allowing endolysins access to the peptidoglycan (PG) cell wall, which they degrade to weaken the cell envelope and promote osmotic lysis. Phages that infect Proteobacteria also encode a spanin in their lysis cassette that functions to disrupt the host outer membrane. The spanin-requirement for cell lysis provided the first clue that the proteobacterial outer membrane confers mechanical rigidity to the cell envelope. Corynebacteria and mycobacteria also build an outer membrane, but it is made of mycolic acids instead of lipopolysaccharides. Here, we investigated whether the mycomembrane presents a mechanical barrier to phage-induced lysis of corynebacteria. In addition to annotated holin and endolysin genes, mycobacteriophages and corynephages were found to encode a membrane protein we call LysZ in their lysis cassettes. Deletion of lysZ in the corynephage Cog blocked lysis of its host Corynebacterium glutamicum. Surprisingly, disruption of the host mycomembrane did not correct this phenotype. Instead, a genetic analysis revealed that blocking synthesis of membrane-anchored glycopolymers called lipomannans/lipoarabinomannans (LM/LAMs) can restore plaque formation when LysZ is inactivated. This genetic system also identified the likely flippase that transports decaprenyl-linked mannose units to the extracellular side of the membrane for polymerization into LM/LAM. Overall, our results indicate that lipoglycans like LM/LAMs play roles in mechanically stabilizing bacterial envelopes and that phages use LysZ-like factors to overcome this barrier to promote lysis.

|
Scooped by
?
June 26, 12:38 AM
|
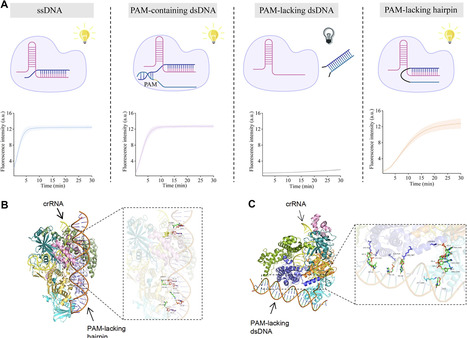
CRISPR–Cas12a has been demonstrated to be activated for its trans-cleavage activity by single- and double-stranded DNA containing a protospacer adjacent motif (PAM), but other types of activators have remained undiscovered. In this work, we found that a hairpin-structured substrate can activate the trans-cleavage activity of Cas12a without a PAM, and the parameters of the hairpin loop obviously affect the activity. Cas12a exhibits sequence preference for proximal loops, preferring to recognize polyadenine hairpin loop activators. Molecular docking and dynamic calculations provide a theoretical basis for the activation of Cas12a by hairpin activators. Leveraging the efficient activation capability of the hairpin activator, we constructed an allosteric detection platform for non-nucleic acid targets, capable of sensitively and specifically detecting hypochlorous acid and calcium ions. This novel activator of Cas12a holds enormous potential for the development of multi-functional biological platforms.

|
Scooped by
?
June 25, 11:47 PM
|
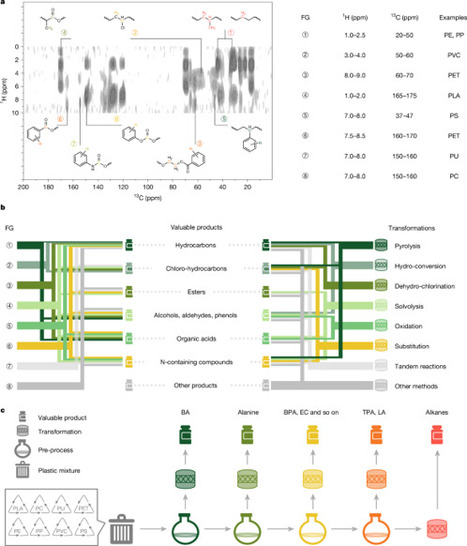
The global crisis of plastic waste accumulation threatens wildlife and ecosystems. Catalytic processes that convert plastic waste into valuable chemicals and fuels offer promising solutions. Recycling or upcycling of real-life plastic mixtures is challenging owing to their diverse composition and structure. Here we propose a product-oriented strategy leveraging the orthogonality in reactivities of different functional groups in plastic mixtures to yield valuable products. This approach involves identifying functional groups followed by converting a selective component in the mixture to valuable products. We use mixtures of polystyrene, polylactic acid, polyurethane, polycarbonate, polyvinyl chloride, polyethylene terephthalate, polyethylene and polypropylene, as well as real-life plastics, to demonstrate the feasibility and effectiveness of the proposed strategy. The diverse physical and chemical properties of these components, which typically hinder direct recovery, offer opportunities for extraction and transformation with the proposed strategy. From a 20-g mixture of real-life plastics, including polystyrene foam, a polylactic acid straw, a polyurethane tube, a polycarbonate mask, a polyvinyl chloride bag, a polyethylene terephthalate bottle, a polyethylene dropper and a polypropylene bottle, we obtained more than 8 separate chemicals: 1.3 g of benzoic acid, 0.5 g of plasticizer, 0.7 g of alanine, 0.7 g of lactic acid, 1.4 g of aromatic amine salt, 2.1 g of bisphenol A, 2.0 g of terephthalic acid and 3.5 g of C3–C6 alkanes. This study reveals the potential for designing transformation strategies for complex plastic waste based on their chemical nature and opens paths for managing end-of-life plastic mixtures. A product-oriented strategy that leverages the reactivities of different functional groups in real-life plastic mixtures can be used to obtain valuable products, opening a path for managing end-of-life plastic mixtures.
|

|
Scooped by
?
June 26, 11:47 PM
|
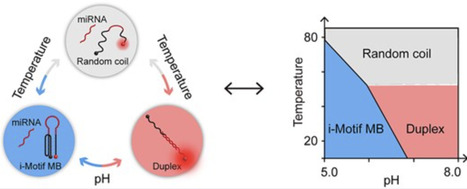
A molecular beacon (MB) is an oligonucleotide probe with a stem-loop structure designed to fluoresce upon hybridization with a specific target nucleic acid. Herein, we introduce a novel i-motif DNA-based molecular beacon (iMB) designed for nucleic acid detection. DNA chemical sequencing showed that the iMB can be selectively opened when its target is present, thanks to the formation of a duplex between this target and the iMB recognition loop. Phase diagrams were introduced to analyze the binding properties between the iMB and its target, and to optimize the detection conditions with different combinations of pH and temperature. Benefiting from the environment-sensitive stability and special structural feature inherent to the i-motif, this iMB exhibits a controllable sensitivity and specificity through choosing appropriate temperature/pH combination and recognition loop position in the i-motif probe. Leveraging the duplex-specific nuclease (DSN) resistance of the i-motif, we significantly enhanced the sensitivity by more than four orders of magnitude through the implementation of a DSN-assisted amplification strategy. Finally, miRNAs in cell lines and patient plasma were detected by the iMBs, as validated via quantitative reverse transcription polymerase chain reaction (RT-qPCR). This research successfully broadens the structural repertoire of MB stems to include a pH- and temperature-responsive i-motif stem.

|
Scooped by
?
June 26, 11:33 PM
|
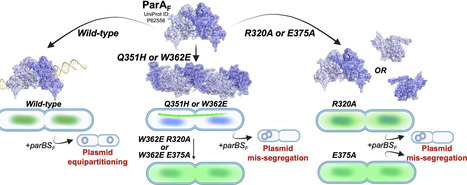
Mobile genetic elements such as plasmids play a crucial role in the emergence of antimicrobial resistance. Hence, plasmid maintenance proteins such as ParA of the Walker A-type ATPases/ParA superfamily are potential targets for novel antibiotics. Plasmid partitioning by ParA relies on ATP-dependent dimerization and formation of chemophoretic gradients of ParA-ATP on bacterial nucleoids. Though polymerization of ParA has been reported in many instances, the need for polymerization in plasmid maintenance remains unclear. In this study, we provide insights into the polymerization of ParA and the effect of polymerization on plasmid maintenance. We report two mutations, Q351H and W362E, in ParA from the F plasmid (ParAF) that form cytoplasmic filaments independent of the ParBSF partitioning complex. Both variants fail to partition plasmids, do not bind non-specific DNA, and act as super-repressors to suppress transcription from the ParAF promoter. Further, we show that the polymerization of ParAF requires an ATP-dependent conformational switch. We identify two residues, R320 in helix 12 and E375 in helix 14 at the interface of the predicted ParAF filament structure, whose mutations abolish filament assembly of ParAF W362E and affect plasmid partitioning. Our results thus suggest a role for the C-terminal helix of ParAF in plasmid maintenance and assembly into higher order structures.

|
Scooped by
?
June 26, 5:50 PM
|
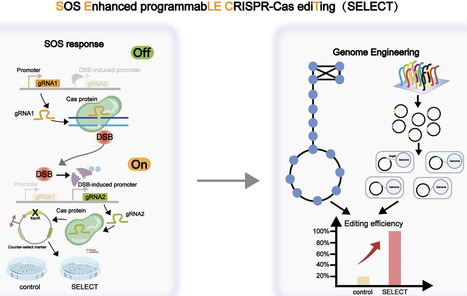
CRISPR-based methods enable genome modifications for diverse applications but often face challenges, such as inconsistent efficiencies, reduced performance in iterative modifications, and difficulties generating high-quality datasets for high-throughput genome engineering. Here, we present SELECT (SOS Enhanced programmabLE CRISPR-Cas ediTing), a novel strategy integrating the CRISPR–Cas system with the DNA damage response. By employing designed and optimized double-strand break induced promoters that are activated upon genome editing, SELECT enables a counter-selection process to eliminate unedited cells, ensuring high-fidelity editing. This approach achieves up to 100% efficiency for point mutations, iterative knockouts, and insertions. In high-throughput library editing, SELECT achieved up to 94.2% efficiency and preserved higher library diversity compared with conventional methods. Application of SELECT in flaviolin biosynthesis resulted in a 3.97-fold increase in production. Furthermore, integration with machine learning tools allowed rapid mapping of genotype–phenotype relationships. SELECT provides a versatile platform for precision genome engineering in Escherichia coli and Saccharomyces cerevisiae.

|
Scooped by
?
June 26, 1:40 AM
|
Escherichia coli lysate-based cell-free systems have gained traction for a variety of point-of-use biological applications. Lysate-based cell-free reactions can be freeze-dried, deployed without requiring cold chain, and have a high ease of use through simple rehydration. To maximize their potential, it is of interest to stabilize these reactions to withstand a variety of conditions for long-term storage and use, including stabilization to UV exposure. To address this issue and aid in point-of-use applications, we investigate the use of synthetic melanin nanoparticles as UV-protective additives that are compatible with cell-free reactions. These particles have broadband absorption properties and radical scavenging activity that allow for protection from free radical generation during prolonged UV exposure. Stabilizing cell-free reactions in this way may prolong the stability for use in the field where exposure to sunlight is inevitable.

|
Scooped by
?
June 26, 1:36 AM
|
The 16S-23S-5S ribosomal RNA (rRNA) gene operon is a cornerstone of ribosomal architecture and function, making it an indispensable molecular marker for microbial taxonomy and phylogenetics. Despite the extensive development of 16S rRNA gene databases, a significant void persists in comprehensive resources that integrate all three rRNA genes (16S, 23S, and 5S), their precise alignments, intragenomic heterogeneity, and intricate structural details. To address this critical research gap, we introduce the 16S-23S-5S rRNA Database, a meticulously curated and intuitive platform encompassing around 5000 complete bacterial and archaeal genomes. This database provides granular operon-level annotations, robust alignment data, quantitative copy number statistics, percent identity matrices, and predicted secondary structures for each rRNA component. Our rigorous methodology integrates covariance models from RFAM with BLAST-based analyses for sequence comparisons, Clustal Omega for multiple sequence alignment, and Circos for genomic visualization, thereby elucidating the genomic architecture, sequence diversity, and structural conservation of rRNA genes. Crucially, the database systematically accounts for intragenomic heterogeneity, a factor paramount for accurate microbial classification and the inference of evolutionary trajectories. Our analysis reveals prevalent phyla such as Pseudomonadota (1,891 genomes), Actinomycetota (846 genomes), and Bacillota (773 genomes), with Bacillota exhibiting the highest average rRNA gene copy numbers. By integrating these comprehensive elements, the 16S-23S-5S rRNA Database not only fills a substantial lacuna in the field of rRNA data analysis but also establishes a foundational resource for large-scale investigations in microbial systematics, genome evolution, and ribosomal biology, benefiting taxonomists, microbial genomics, and microbiome researchers alike.

|
Scooped by
?
June 26, 1:32 AM
|
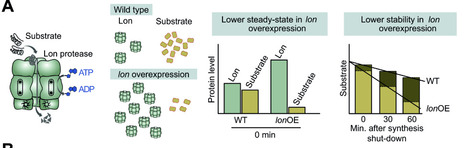
The ATP-dependent cytoplasmic protease Lon has critical functions in protein quality control and cellular regulation in organisms across the three domains of life. In the opportunistic pathogen Pseudomonas aeruginosa, lon loss-of-function mutants exhibit multiple phenotypic defects in motility, virulence, antibiotic tolerance and biofilm formation. However, only a couple of native substrate proteins of Lon are described in P. aeruginosa until now and most of the phenotypes associated with Lon remain unexplained. Here, we searched for novel Lon substrates in P. aeruginosa by analyzing proteome-wide changes in protein levels and stabilities following lon overexpression. Our search yielded a large number of putative Lon substrates with diverse cellular functions, including metabolic enzymes, stress proteins and a significant fraction of motility-related proteins. In vitro degradation assays confirmed the metabolic protein SpeH, the heat shock protein IbpA as well as seven proteins involved in flagella- and type IV pilus-mediated motility as novel substrates of Lon. The new motility-associated substrates include both key regulators of motility (FliA, RpoN, AmrZ) as well as structural flagellar components (FliG, FliS and FlgE). Further, by isolating suppressor mutations bypassing the motility defect of lon- cells, we reveal that Lon-dependent degradation of the specific substrate SulA, a cell division inhibitor, is crucial for ensuring proper cell division and motility under optimal conditions. In sum, our work highlights Lon’s regulatory role in degrading functional proteins involved in critical cellular processes and contributes to a better molecular understanding of the pathways underlying Pseudomonas pathogenicity.

|
Scooped by
?
June 26, 1:24 AM
|
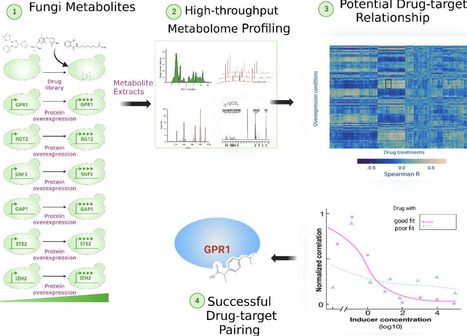
Metabolomics, a critical tool for analyzing small-molecule metabolites, integrates with genomics, transcriptomics, and proteomics to provide a systems-level understanding of fungal biology. By mapping metabolic networks, it elucidates regulatory mechanisms driving physiological and ecological adaptations. In fungal pathogenesis, metabolomics reveals host-pathogen dynamics, identifying virulence factors like gliotoxin in Aspergillus fumigatus and metabolic shifts, such as glyoxylate cycle upregulation in Candida albicans. Ecologically, it highlights fungal responses to abiotic stressors, including osmolyte production like trehalose, enhancing survival in extreme environments. These insights highlight metabolomics’ role in decoding fungal persistence and niche colonization. In drug discovery, it aids target identification by profiling biosynthetic pathways, supporting novel antifungal and nanostructured therapy development. Combined with multi-omics, metabolomics advances insights into fungal pathogenesis, ecological interactions, and therapeutic innovation, offering translational potential for addressing antifungal resistance and improving treatment outcomes for fungal infections. Its progress shed light on complex fungal molecular profiles, advancing discovery and innovation in fungal biology.

|
Scooped by
?
June 26, 1:15 AM
|
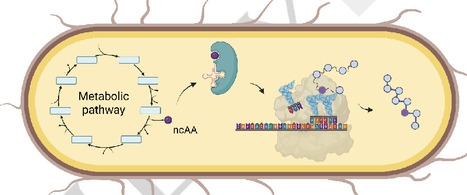
Autonomous cells are engineered biological systems capable of biosynthesizing and directly incorporating non-canonical amino acids (ncAAs) into proteins. These systems have the potential to extend the applicability of the genetic code to enable large-scale fermentative production of proteins carrying ncAAs. This work evaluates approaches for the generation of autonomous and semi-autonomous cells. Semi-autonomous cells rely on the external addition of a precursor, which is enzymatically converted in vivo to an ncAA that is directly incorporated. In contrast, autonomous cells have a metabolic system that produces and directly incorporates an ncAA in vivo. Through a critical evaluation of the state of the art, the reader is provided with an opinion on the future development of the field.

|
Scooped by
?
June 26, 1:03 AM
|
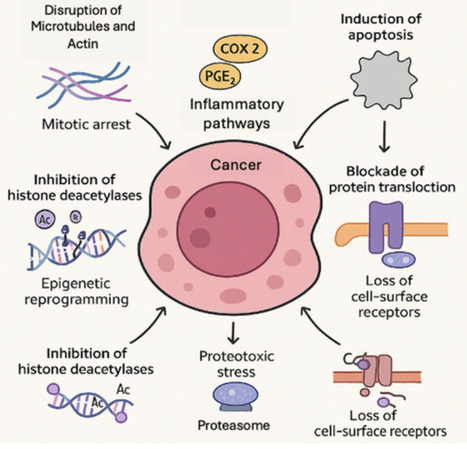
Cyanobacteria-derived peptides represent a promising class of anti-cancer agents due to their structural diversity and potent bioactivity. They exert cytotoxic effects through mechanisms including microtubule disruption, histone deacetylase inhibition, and apoptosis induction. Several peptides—most notably the dolastatin-derived auristatins—have achieved clinical success as cytotoxic payloads in antibody–drug conjugates (ADCs). However, challenges such as limited tumor selectivity, systemic toxicity, and production scalability remain barriers to broader application. Recent advances in targeted delivery technologies, combination therapy strategies, synthetic biology, and genome mining offer promising solutions. Emerging data from preclinical and clinical studies highlight their therapeutic potential, particularly in treatment-resistant cancers. In this review, we (i) summarize key cyanobacterial peptides and their molecular mechanisms of action, (ii) examine progress toward clinical translation, and (iii) explore biotechnological approaches enabling sustainable production and structural diversification. We also discuss future directions for enhancing specificity and the therapeutic index to fully exploit the potential of these marine-derived peptides in oncology.

|
Scooped by
?
June 26, 12:46 AM
|
Global agriculture stands at a critical juncture, facing the dual challenge of sustaining food production for a rapidly growing population while mitigating the environmental consequences of intensive farming. The overuse of chemical fertilizers and pesticides has accelerated soil degradation, biodiversity loss, and ecological imbalances, threatening long-term viability. Synthetic microbial communities (SynCom) have emerged as a promising approach to reshape plant-microbe interactions, offering a precise, scalable, and ecologically sustainable alternative to conventional agrochemicals. Unlike native microbial communities, which form naturally and vary with environmental conditions, SynComs are deliberately assembled consortium of multiple microbial strains selected for their complementary functions, ecological compatibility, and ability to perform targeted roles within a host or environment. By engineering microbes with targeted functional traits, SynComs enhance nutrient assimilation, bolster plant defence, and fortify resilience against biotic and abiotic stresses. The understanding of SynCom design, exploring their composition, functional dynamics, and mechanisms for optimizing plant health is crucial for effective synthesis and application, alongside cutting-edge computational tools and genomic databases that enable precision engineering of microbial communities. Despite their transformative potential, large-scale application of SynComs remains constrained by challenges related to field efficacy, regulatory frameworks, and long-term microbial persistence. Addressing these barriers through interdisciplinary research and policy innovation is imperative. As environmental microbiome moves towards sustainability-driven solutions, SynComs hold the key to revolutionizing farming practices, reducing chemical dependence, and ensuring global food security in an era of mounting environmental stressors.

|
Scooped by
?
June 26, 12:40 AM
|
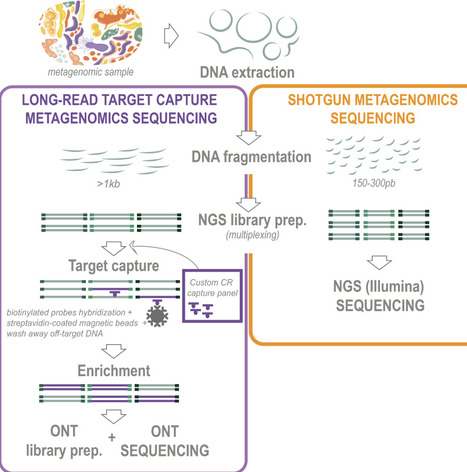
Chemoreception plays a central role in microbial adaptability, influencing both community structure and interactions with the environment. However, many chemosensitive microorganisms occur at low abundances in natural ecosystems, which has limited their detection and study using conventional metagenomic sequencing. Here, we employed a custom-designed target capture sequencing approach—encompassing all known chemoreceptor genes from both cultured and uncultured microorganisms—to uncover and characterize the vast chemosensory potential and biodiversity within the rarest fractions of the microbiome. Compared to standard environmental sequencing methods, our approach enhanced the detection of chemoreceptor (CR) genes and their associated sensing domains by orders of magnitude across diverse environments, including the rhizosphere, phyllosphere, soil, aquatic ecosystems, the human gut, and bovine rumen. This enabled the identification of thousands of low-abundance chemosensitive microorganisms that remained undetectable using conventional sequencing approaches, including known plant pathogens and symbionts. Phylogenetic analysis of the most divergent CR genes revealed evidence for novel chemosensitive species, potentially representing new bacterial phyla and classes. Our study provides a new perspective on the chemoreception capabilities of environmental microbes and opens new avenues for discovering and characterizing novel microbial sensing mechanisms.

|
Scooped by
?
June 26, 12:08 AM
|
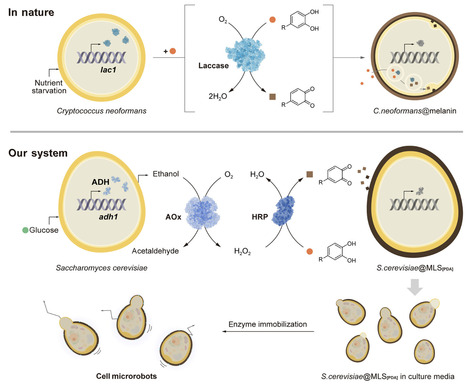
Living organisms use intricate strategies to adapt and survive in response to potentially lethal environment changes. Inspired by cryptobiosis in nature, researchers have pioneered approaches to create cell-in-shell nano-biohybrids, aiming to endow cells with enhanced protection and exogenous functions. Yet, these methods still lack the biological autonomy intrinsic to natural cellular responses. Here, we present an innovative chemo-metabolically coupled strategy for the autonomous construction of cell-in-shell structures in cell growth medium. Our system harnesses ethanol fermentation by Saccharomyces cerevisiae, chemically coupled with an enzymatic cascade involving alcohol oxidase and horseradish peroxidase, to drive the nanoshell formation of polydopamine. The integration of autonomous shell formation with cellular proliferation produces anisotropic cell-in-shell structures, which can serve as enzyme-powered cell microrobots, upon conjugation with urease. Our autonomous system enables the creation of cell-in-shell nanobiohybrids with dynamic and adaptive environmental interactions, paving the way for transformative applications in synthetic biology, such as artificial cells, as well as advancements in cell-based therapies.
|
 Your new post is loading...
Your new post is loading...