Your new post is loading...

|
Scooped by
?
Today, 4:31 PM
|
Predicting changes in protein thermostability caused by amino acid substitutions is essential for understanding human diseases and engineering proteins for practical applications. While recent protein generative models demonstrate impressive zero-shot performance in predicting various protein properties without task-specific training, their strong unsupervised prediction ability remains underexploited to improve protein stability prediction. We present SPURS, a deep learning framework that rewires and integrates two complementary protein generative models–a protein language model and an inverse folding model–and reprograms this unified framework for stability prediction through supervised fine-tuning on mega-scale thermostability data. SPURS delivers accurate, efficient, and scalable stability predictions and generalizes to unseen proteins and mutations. Beyond stability prediction, SPURS enables broad applications in protein informatics, including zero-shot identification of functional residues, improved low-N protein fitness prediction, and systematic dissection of stability-pathogenicity for human diseases. Together, these capabilities establish SPURS as a versatile tool for advancing protein stability prediction and protein engineering at scale. Understanding how mutations alter protein stability is essential for biology and disease research. Here, the authors develop SPURS, a model that rewires pre-trained protein generative models to accurately predict stability changes.

|
Scooped by
?
Today, 4:26 PM
|
Tumor-targeted bacteria have emerged as promising drug carriers due to their intrinsic motility and hypoxia-homing property. Therapeutic agents can be loaded onto the bacterial surface, enabling their active delivery into tumor tissues. However, premature drug release during systemic circulation—likely triggered by various physiological/physical factors—inevitably results in reduced efficacy or increased off-target toxicity. Here, we present a genetic engineering strategy that enables E. coli MG1655 (EC) to autonomously produce a biofilm “jacket” on its surface (termed MEC) by regulating the expression of the biofilm-associated Csg gene cluster. This biofilm coating markedly enhances drug adsorption (1.7-fold increase for the model drug indocyanine green, ICG) and effectively prevents off-target leakage during systemic circulation. Benefiting from its tumor-homing capability and biofilm-mediated protection, MEC can deliver substantially more ICG into tumor inner regions. In murine tumor models, MEC-mediated delivery achieves significantly enhanced intratumoral drug retention and photothermal efficacy in comparison with the wild-type bacterial carrier. This work demonstrates an effective tumor-targeted drug delivery strategy based on genetically engineered biofilm technology, offering a promising avenue for precision bacterial oncology.

|
Scooped by
?
Today, 4:15 PM
|
Heme, a protoporphyrin IX iron complex, functions as an essential prosthetic group in hemoglobin and myoglobin, mediating oxygen storage and transport. Additionally, heme serves as a critical cofactor in various enzymes such as cytochrome c, enabling electron transfer within the mitochondrial respiratory chain. Unlike protein-bound heme, free or labile heme exhibits cytotoxic, pro-oxidant, and pro-inflammatory properties. Elevated levels of free heme are associated with various pathophysiological conditions, including hemolytic disorders such as sickle cell disease, malaria, and sepsis. In this review, we introduce the physiological roles of heme and its involvement in human health and disease. We also examine the mechanisms of heme sensing and regulation in bacterial cells. A variety of analytical methods have been developed to detect and quantify heme, enabling differentiation between protein-bound and free forms. These tools are discussed in the context of their applications in studying cellular heme regulation and their use in monitoring pathological conditions in humans. In particular, we describe examples of biosensors employing bacterial heme sensor proteins as recognition elements.

|
Scooped by
?
Today, 4:10 PM
|
Bacteriophages are major drivers of bacterial population dynamics, yet the significance of post-transcriptional regulation during infection remains largely unexplored. Central to this regulatory layer are small RNAs (sRNAs), which regulate target mRNAs via base-pairing, typically facilitated by RNA chaperones such as Hfq. Here, we applied RNA interaction by ligation and sequencing (RIL-seq) to comprehensively map the in vivo RNA-RNA interaction network in E. coli during phage lambda infection. This analysis revealed extensive reprogramming of E. coli-E. coli interactions, phage-specific lambda-lambda interactions, and interkingdom interactions between phage and host RNAs. Among these, we identified a phage-encoded sRNA, phage replication enhancer sRNA (PreS), embedded within the early left operon. PreS regulates essential host genes, including dnaN, which encodes the DNA polymerase β sliding clamp. This regulation enhances DNA replication and fine-tunes the phage lytic cycle. These findings uncover an RNA-level regulatory layer in phage-host interactions and demonstrate how a phage-encoded sRNA can hijack host replication machinery to optimize infection.

|
Scooped by
?
Today, 3:58 PM
|
CRISPR–Cas systems have revolutionized genome engineering technologies, but type IV CRISPR–Cas systems and their genome engineering potential have been critically underexplored. In this study, we identified a type IV-A3 CRISPR–Cas system from a clinical Klebsiella pneumoniae isolate and characterized its plasmid targeting activity and capacity to suppress chromosomal and plasmid gene expression in E. coli. We revealed the pivotal role of Csf3 (Cas5) and the dispensable roles of Csf1 (Cas8-like) and Csf4 (DinG helicase) subunits in IV-A3 CRISPR–Cas complex formation. The system prevents plasmid propagation via interplay between DinG helicase activity and strategic protospacer positioning relative to plasmid replication and maintenance components. We enabled the IV-A3 CRISPR–Cas system to introduce lethal, sequence-specific double-stranded (ds)DNA breaks in the E. coli chromosome by fusing the nuclease domain of the I-TevI nuclease to the Cas8 N-terminus. Further, we developed a series of base editors, with various editing efficiencies and windows, by fusing the PmCDA1 cytidine deaminase to the Cas8, Cas5, and DinG subunits. Finally, conjugative transfer of the Cas5–PmCDA1 base editor into E. coli deactivated the tryptophan repressor gene, boosting IAA production. Our study provides new insights into type IV-A3 CRISPR–Cas systems and highlights their potential in genome engineering applications.

|
Scooped by
?
Today, 3:50 PM
|
CRISPR–based environmental biosurveillance (CRISPR-eBx) offers a portable, specific, sensitive, and cost-effective platform for detecting organisms from environmental nucleic acids. Applications are broad, ranging from pathogen detection to monitoring invasive and endangered species across a range of environmental sources, including water, soil, and air. However, if CRISPR-eBx is to be deployed for novel biological/gene targets and environmental sources, key challenges must be addressed. This review synthesizes recent developments at the intersection of CRISPR technology, computational science, synthetic biology, and biosurveillance. We highlight promising innovations and identify knowledge gaps to present a strategic road map for establishing CRISPR-eBx as a next-generation, frontline biosurveillance solution.

|
Scooped by
?
Today, 3:42 PM
|
Cross-kingdom RNA interference (ck-RNAi) is a biological process in which small RNA (sRNA) molecules are transferred between organisms belonging to different kingdoms to silence specific genes. Although numerous instances of reciprocal ck-RNAi have been documented in plants, demonstrating a modulation of the interaction between plants and their pathogens, pests, or symbiotic partners, the underlying molecular mechanisms remain largely elusive. In this article, we distinguish between naturally occurring and transgene-based cases of ck-RNAi, examine the diverse mechanisms governing the transfer of primary ck-RNAi signals from donor to recipient organisms, and explore the prerequisites for their amplification and systemic spread. Finally, we highlight key unresolved questions concerning the mechanistic basis of ck-RNAi and offer a perspective on its potential role in co-evolutionary dynamics.

|
Scooped by
?
Today, 2:12 PM
|
The cellular context interacts with genetic circuits, decisively defining their performance. However, contextual dependencies (the interplay between the host and the circuit) are often difficult to engineer rationally, leading to a lack of control over circuit behavior. To address this challenge, we replaced the native regulatory machinery of the RNA polymerase (RNAP), the cell's core protein-making machine, from the bacteria Pseudomonas putida KT2440 with an inducible system, enabling tunable growth regulation and thereby gaining systemic control over the entire cellular machinery. Specifically, this was achieved by placing key components, the β and β' subunits of the enzyme, under the control of the XylS-Pm inducible system. By using its cognate chemical inducer, 3-methylbenzoate, cell's growth can be controlled at will, enabling the precise tuning of the cellular context into distinct, stable states. We correlated genetic circuit behavior with the cell's growth state by observing the constitutive expression of a reporter gene and the performance of a collection of genetic NOT logic gates. Our results show that the modulation of contextual dependencies is specific to the circuit components but not a random process. A mathematical model allowed us to classify that modulation into three different categories for our library of NOT gates, based on the prediction of how RNAP availability shapes host-circuit interaction. Finally, using growth control as an input for a 2-input circuit lead to a NAND gate with the potential for morphological computing, where the cell's physical body itself undertakes part of the information processing. Our findings indicate that growth control can be used as an engineering parameter, allowing us to search for optimal scenarios that enhance the potential of genetic tools.

|
Scooped by
?
Today, 12:54 PM
|
Driven by the persistence of microplastics and an overdependence on non-renewable sources, mycelium materials have emerged as alternatives to traditional materials, owing to their sustainable production and versatility. Engineered living materials, composite material systems incorporating biological components to enable function, possess desirable regenerative properties but have yet to be fully applied due to their lack of strength versus synthetic or natural materials. We leverage two strategies used by nature to generate mechanical strength: structural hierarchy and biomineralization. We extrude a bioink composed of alginate and a modified strain of the fungus Aspergillus niger capable of silica mineralization, and modulate the morphology by the growth and processing conditions. Mineralization results in significantly stronger and stiffer dried fibers. We demonstrate their potential as textile materials through twisting and braiding to significantly increase the fracture strain. Our results show that mycelium morphology and mechanics can be tuned through mineralization, growth conditions, and processing.

|
Scooped by
?
Today, 11:29 AM
|
Bacterial extracellular vesicles (BEVs) are known to enhance infection susceptibility in vivo, yet the mechanistic basis for this remote preconditioning of host cells is unknown. Here, we discover an evolutionarily conserved, lipid-driven physical mechanism by which pathogenic bacterial EVs systemically arrest phagosome maturation in bystander host cells. Using live-cell fluorescence lifetime imaging, in vitro reconstitution and micromanipulation we show that EVs from diverse pathogens - Mycobacterium tuberculosis, Klebsiella pneumoniae, and Staphylococcus aureus - fuse with host plasma, phagosomal, and lysosomal membranes. This fusion increases membrane tension and perturbs early phagosomal maturation. Transcriptomic profiling confirms a broad downregulation of phagosome maturation genes while upregulation of lysosomal stress responsive genes. Crucially, in vitro reconstitution shows that EVs, and their purified lipids alone, are sufficient to induce phase separation and increase membrane tension, directly inhibiting phago-lysosomal fusion. Our findings establish a paradigm in which pathogens exploit EVs not merely as delivery vehicles, but as tools to remotely rewire host cell membrane mechanics to hijack phagosome maturation and host defense - a strategy that moves beyond canonical effector-based models of pathogenesis.

|
Scooped by
?
Today, 11:13 AM
|
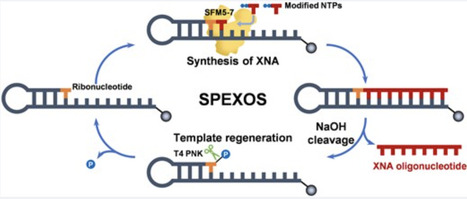
DNA-based information storage offers a promising alternative to conventional media due to its high density, long-term stability, and low energy requirements. However, its application remains hindered by synthesis costs, limited sequence length and poor scalability. DNA polymerase is a critical enzymatic tool in the DNA storage systems by enabling high-fidelity data writing and targeted sequence amplification. In this study, we engineered chimeric DNA polymerases by fusing the high-fidelity 9°N DNA polymerase with double-stranded DNA binding proteins derived from thermophilic archaea. These fusions significantly enhanced processivity, thermal stability, and salt tolerance by stabilizing enzyme-template interactions, mimicking sliding clamps while preserving catalytic efficiency. Leveraging these properties, we demonstrated precise file retrieval from a mixed oligonucleotide pool using orthogonal barcode primers. Compared with wild-type 9°N, the chimeric polymerases, particularly PLS, exhibited reduced substitution error rates and improved read accuracy. We then applied these enzymes to a DNA movable type storage system, where prefabricated DNA modules were assembled into encoding blocks. Using engineered polymerases, these blocks were recombined to enable flexible data rewriting without de novo DNA synthesis. Moreover, a multi-enzyme assembly strategy enabled the construction of kilobase-scale DNA sequences encoding a classical Chinese poem, achieving complete data recovery. All assembled fragments remained stable in E. coli over 100 generations, exhibiting the potential for in vivo storage. Collectively, our findings demonstrated the role of engineered DNA polymerases for DNA-based information storage. Moreover, this system reduced synthesis demands, supported scalable rewriting, and ensured long-term preservation, offering a practical route to sustainable, high-fidelity DNA data storage.

|
Scooped by
?
December 19, 4:57 PM
|
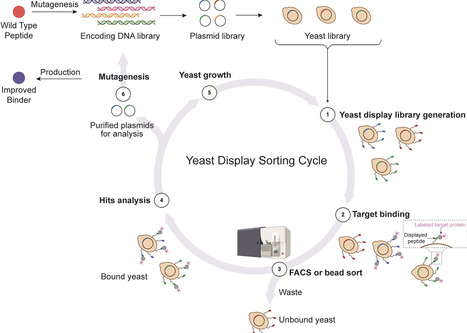
Cyclic disulfide-rich peptides have become increasingly popular in drug development because their structures enhance molecular stability and allow for mutagenesis to introduce non-native functions. This review focuses on yeast-based platform technologies and their utility in advancing cyclic disulfide-rich peptides as drug modalities and for large-scale biomanufacturing. These technologies include yeast surface display which facilitates the screening of large libraries to develop peptide binders with strong affinity and selectivity for protein targets, while maintaining the innate high stability of the peptide scaffold via protease-based selection pressure. We also describe a recently developed platform that leverages yeast’s ability to secrete correctly folded disulfide-rich peptides while simultaneously displaying peptide or protein tags on their surfaces. In combination with microfluidics technology, the platform creates single-cell yeast-in-droplets reactors, enabling the screening of large libraries based on functional output rather than solely on binding affinity. After identifying cyclic peptide candidates through library-based discovery, these candidates can be produced using a versatile yeast-based bioproduction platform. Traditionally, cyclic disulfide-rich peptides are produced through solid-phase synthesis, a method that generates significant amounts of toxic waste. In contrast, yeast-based bioproduction offers an environmentally sustainable alternative. It has the capability to produce structurally distinct peptides with minimal adjustments and is easily scalable using microbial fermenters, making it an ideal choice for large-scale production.

|
Scooped by
?
December 19, 4:41 PM
|
Tissue- or cell-type-specific expression of transgenes is often essential for interrogation of the biological phenomenon or predictable engineering of multicellular organisms but can be stymied by cryptic enhancers that make identification of promoters that generate desired expression profiles challenging. In plants, the months-to-years long timeline associated with prototyping putative tissue-specific promoters in transgenic lines deepens this challenge. We have developed a novel strategy called Ribozyme-Enabled Tissue Specificity (RETS) that leverages the knowledge of where and when genes are expressed derived from transcriptomic studies to enable tissue-specific expression without needing characterized promoters. It uses a split self-splicing ribozyme based on a group I intron from Tetrahymena thermophila to enable the conditional reconstitution of a transgene mRNA in the presence of a secondary tissue-specific mRNA of choice. We elucidate the design features that enable flexible swapping of transgenes and targets, enhancing transgene expression, and circumventing host RNA interference responses. We then show that these innovations enable tissue-specific and dose-dependent expression of transgenes in Arabidopsis thaliana. Finally, we demonstrate the utility of RETS both for creating genetically encoded biosensors to study the spatiotemporal patterns of gene expression in planta and for engineering tissue-specific changes in organ size. RETS provides a novel avenue to study expression patterns of native loci with nondestructive imaging, complementing the weakness of existing approaches. Additionally, the spatiotemporal control of transgene expression afforded by RETS enables precision engineering of plant phenotypes, which will facilitate enhancing crops without the trade-offs associated with constitutive expression.
|

|
Scooped by
?
Today, 4:29 PM
|
The production of recombinant proteins in E. coli is often hampered by the formation of inclusion bodies. While fusion tags can enhance solubility, existing systems are hampered by a lack of standardization, with tags scattered across disparate plasmid backbones and inconsistent cloning sites, complicating parallel screening. To address this, we constructed a standardized series of expression vectors, termed pNX, by incorporating nine small fusion tags (SUMO, LD, ACP, BCCP, GB1, Fh8, SmbP, TolA, and TrxA) into a uniform pET-28b backbone. Each pNX vector features an identical configuration: a T7 promoter, an N-terminal fusion tag, a synthetic linker, a TEV protease cleavage site, and a multiple cloning site (MCS) flanked by dual 6×His tags. We evaluated this system using four model proteins (EcFabG, eGFP, XccXanA2, and XccXanL). Our results showed that specific tags significantly improved both the expression level and solubility of the target proteins without compromising their biological activity. Notably, the lipoyl domain (LD) was identified, to our knowledge for the first time, as an effective solubility enhancer. The standardized MCS enabled rapid, parallel cloning, facilitating the efficient screening of optimal fusion partners. The pNX vector series provides a versatile and efficient platform for enhancing the soluble expression of challenging recombinant proteins in E. coli, streamlining the empirical identification of ideal fusion tags.

|
Scooped by
?
Today, 4:24 PM
|
The discovery of the antiviral-responsive RNA editing system is a significant advance in eukaryotic biology. It reveals new dimensions of RNA editing diversity and fungal antiviral strategies, and provides compelling evidence for the adaptive significance of RNA editing in host-virus coevolution. This discovery establishes a novel model for investigating both the molecular mechanisms of RNA editing and the evolutionary dynamics of altruistic defense.

|
Scooped by
?
Today, 4:12 PM
|
Extracellular vesicles (EVs) are lipid-bound nanocarriers released by various eukaryotic cells and found in diverse bodily fluids. EVs have transitioned from being considered cellular waste disposers to significant players in intercellular communication and signaling. These EVs carry signature cargos of infected cells and thus can be helpful as biomarkers or prognostic markers for infectious diseases. Viruses can manipulate the EV biogenesis machinery in their own dissemination. EVs released from virus-infected cells can carry immune modulatory molecules, thus contributing to disease progression. This comprehensive review collates the information on the impact of EVs on viral infection and disease progression.

|
Scooped by
?
Today, 4:00 PM
|
Antimicrobial resistance (AMR) presents a mounting global health crisis. Traditional antibiotic discovery methods are hindered by low throughput, frequent rediscovery of known compounds, and limited access to unculturable microbes and silent biosynthetic gene clusters (BGCs). Droplet microfluidics offers a transformative platform to overcome these barriers by enabling massively parallel, compartmentalized analysis at the single-cell level. In this perspective, we discuss how droplet microfluidics is reshaping the antibiotic discovery landscape by addressing long-standing limitations in cultivation, screening, and compound identification. We highlight recent advances in: (1) accessing microbial dark matter through high-throughput single-cell technologies, (2) activating cryptic BGCs and dissecting microbial interactions using microfluidic coculture platforms, (3) accelerating chemical dereplication and discovery of novel metabolites through integration with mass spectrometry-based tools, and (4) uncovering resistance and persistence mechanisms by linking antibiotic resistance genes to individual microbial hosts and single cell transcriptomes. Together, these innovations position droplet microfluidics as a powerful engine to accelerate antibiotic discovery and combat the global threat of AMR.

|
Scooped by
?
Today, 3:53 PM
|
Biologically meaningful interpretation of transcriptomic datasets remains challenging, particularly when context-specific gene sets are either unavailable or too generic to capture the underlying biology. We here present InCURA, an integrative clustering strategy based on transcription factor (TF) motif occurrence patterns in gene promoters. InCURA takes as input lists of (i) all expressed genes, used solely to identify dataset-specific expressed TFs, and (ii) differentially regulated genes (DRGs) used for clustering. Promoter sequences of DRGs are scanned for TF binding motifs, and the resulting counts are compiled into a gene-by-TFBS matrix. InCURA then uses unsupervised clustering to infer gene modules with shared predicted regulatory input. Applying InCURA to diverse biological datasets, we uncovered functionally coherent gene modules revealing upstream regulators and regulatory programs that standard enrichment or co-expression analyses fail to detect. In summary, InCURA provides a user-friendly, regulation-centric tool for dissecting transcriptional responses, particularly in settings lacking context-specific gene sets.

|
Scooped by
?
Today, 3:44 PM
|
Cell sorting, essential for diagnostics and early intervention, has evolved from conventional methods to sophisticated microfluidic approaches. These miniaturized systems leverage precise hydrodynamic control, facilitating major advances in tumor cell isolation, single-cell analysis, and biomarker detection. However, the vast imaging data generated by these microfluidic techniques necessitate advanced computational methods. Machine learning, particularly computer vision and deep learning, now offers transformative capabilities for automated feature extraction, pattern recognition, and real-time classification, enhancing sorting accuracy, accelerating diagnostics, and informing clinical decisions. This review synthesizes the convergence of microfluidics and machine intelligence, examining their synergistic roles in flow-field optimization, cellular classification, and error correction. While highlighting breakthroughs in diagnostic sensitivity and analytical throughput, we critically address challenges including model generalizability and hardware-software integration. Last, we provide an outlook on multimodal data fusion and the development of on-chip intelligent systems, proposing a roadmap for advancing precision medicine through embedded, adaptive biosensing platforms.

|
Scooped by
?
Today, 2:47 PM
|
Tumor immunotherapy has garnered significant attention, however, several notable challenges remain to be addressed, including: 1) enhancing active targeting, 2) effectively shielding immune checkpoints, and 3) inducing the transformation of “cold” tumor into “hot”. In this study, an “all-in-one” extracellular anaerobic bacterial nanocomposite system is constructed. Escherichia coli Nissle 1917 (EcN) is enveloped with polydopamine via self-polymerization (PDAEcN), subsequently conjugated with a chitosan oligosaccharide (COS) nanoparticle immunostimulant backpack, denoted as PDAEcN/COS. The PDA coating is capable of concealing EcN polysaccharides, masking the immunogenic bacterial surface antigens, and inducing a mild photothermal therapy (PTT) effect. Moreover, PDAEcN/COS exhibits targeted accumulation in hypoxic regions of solid tumors and demonstrates pronounced enrichment on tumor cell surfaces, attributed to the bacterial hypoxic region targeting ability and adhesive properties of PDA, physically obstructing the immune checkpoint. Simultaneously, both EcN and COS markedly enhanced the transformation of M2 macrophages into the M1 phenotype, whereas mild PTT-induced immunogenic cell death (ICD) further mitigated the immunosuppressive nature of the hypoxic tumor microenvironments (TMEs). This integrated therapeutic approach eradicated tumors without eliciting metastasis or discernible side effects following a single injection and laser irradiation in a murine 4T1 cancer model. Ultimately, the immunostimulatory capacity of PDAEcN/COS is considered to hold significant potential for developing novel and efficacious therapies for immunologically “cold” triple-negative breast cancer (TNBC).

|
Scooped by
?
Today, 1:26 PM
|
Although the α-amylase (AmyS) from Geobacillus stearothermophilus exhibits high thermostability, its low enzymatic activity (44.36 ± 2.02 U/mL) severely hinders industrial applications. Given its strong protein secretion capability and GRAS (generally recognized as safe) status, this study employed Bacillus subtilis as the host to enhance AmyS production. Through error-prone PCR and high-throughput screening, a triple mutant T151A/K178E/T458A (AmySM) was generated, showing a 17.67% increase in activity (51.34 ± 1.11 U/mL). AmySM activity was further increased by 29.32% to 65.23 ± 2.33 U/mL using the signal peptide SPykwD, selected from a comprehensive library for superior secretion efficiency. The promoter PgsiB enhanced activity by 37.61% to 89.54 ± 2.95 U/mL. Optimizing the ribosome binding site (RBS) resulted in an additional 48.83% increase in activity, yielding a final activity of 134.02 ± 3.54 U/mL, which corresponds to a 3.02-fold improvement over the initial strain WBSW (44.36 ± 2.02 U/mL). Ultimately, scale-up fermentation in a 5-L bioreactor yielded a maximum extracellular activity of 1244.17 ± 48.66 U/mL at 72 hours, a 2.84-fold increase over the control. This multi-level strategy provides a rational framework for high-efficiency AmySM production, paving the way for extracellular production of high-value proteins in the GRAS host B. subtilis.

|
Scooped by
?
Today, 12:45 PM
|
Recent advances in AI have substantially accelerated in silico drug discovery, yet most computational approaches remain task-specific and require expert knowledge. We present BioChemAIgent, an agentic framework that orchestrates state-of-the-art AI models and established computational chemistry tools to support end-to-end small-molecule analysis, protein modeling, molecular docking, and interaction analysis through a unified interface. BioChemAIgent emphasizes transparent reasoning, reproducible workflows, and community-oriented extensibility for structural biology and drug discovery applications.

|
Scooped by
?
Today, 11:23 AM
|
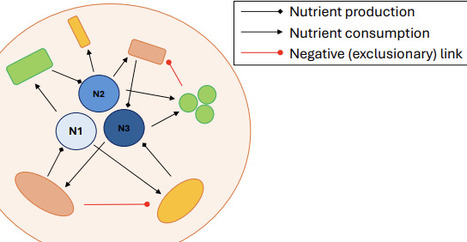
Predicting the species composition of microbial communities is a problem that has proven surprisingly difficult, including in fully controlled microbial systems in a lab setting. Few organisms are able to establish themselves in an community with no others, even if nutrients are provided. At the same time some species are observed to be unable to stably coexist with certain other species. We here present a model attempting to reduce the dynamics of community assembly to its most basic components, while preserving its salient features. The model deals not with abundances or densities of bacterial populations or resources, but only with their presence or absence. Similarly, only three types of discrete relationships between species are considered: species excluding each other, and mutualistic dependence on and production of nutrients in the form of exometabolites. Despite these simplifications, the model system still exhibits rich dynamics, including extinction cascades and emerging stability against invasion. We derive conditions for these to occur and compare our results with existing knowledge of microbial communities. Our results provide a novel approach for the theoretical study of microbial communities and their stability.

|
Scooped by
?
December 19, 5:04 PM
|
Remarkable progress has been achieved by machine learning, particularly in accurate prediction of protein tertiary structures. Despite these advances, accurately annotating protein functions through machine learning approaches remains challenging, primarily due to the limited availability of large-scale benchmarking data. In this study, we addressed this gap by systematically screening proteins from the UniProt database for functional annotations, resulting in the creation of a benchmarking dataset that includes protein sequences and their corresponding annotations. The Protein Annotation Dataset (PAD) is a resource available to train a wide range of machine learning models for assignment of function annotations to previously unlabeled proteins. We curated a comprehensive dataset comprising four categories of functional annotations using enzyme commission (EC) numbers and gene ontology (GO) terms. The dataset was subsequently partitioned into training, validation, and test subsets. Furthermore, we incorporated an independent set from 12 diverse species, enabling the development and evaluation of innovative machine learning models.

|
Scooped by
?
December 19, 4:50 PM
|
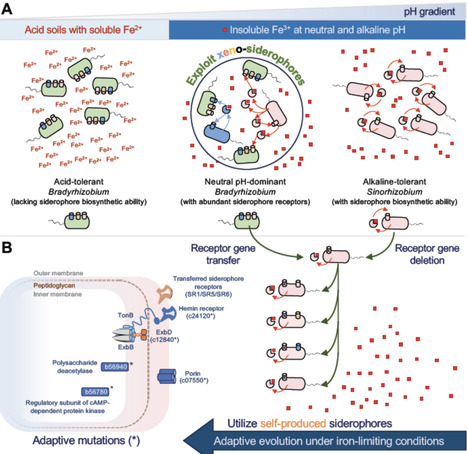
Bradyrhizobium and Sinorhizobium are dominant soybean microsymbionts in acidic/neutral and alkaline soils, respectively. However, the molecular mechanisms underlying this pH-dependent adaptation remain elusive. In this study, phylogenomic analysis of 286 Bradyrhizobium and 322 Sinorhizobium genomes revealed that Bradyrhizobium possesses abundant xeno-siderophore receptors but has limited siderophore biosynthesis functions. In contrast, gene clusters directing siderophore biosynthesis are enriched in Sinorhizobium. As siderophores can chelate the prevalent insoluble Fe3+ under neutral and alkaline conditions, whereas being less important in acidic environments where soluble Fe2+ is readily accessible, we hypothesized that the genus-dependent phyletic distribution of siderophore biosynthesis and exploitation functions may contribute to the pH adaptation of these two genera. Indeed, Bradyrhizobium species barely grow under iron-limiting conditions, and this growth defect can be rescued by xeno-siderophores produced by Sinorhizobium. Using a xeno-siderophore-exploiting Bradyrhizobium diazoefficiens strain, an engineered xeno-siderophore exploiter, and an altruistic siderophore-producing strain derived from Sinorhizobium fredii, we revealed the competitive advantage of xeno-siderophore exploitation during soybean nodulation. Heterologous expression of certain Bradyrhizobium xeno-siderophore receptors, along with various adaptive mutations in the genome of the S. fredii receptor-lacking mutant, allowed this mutant to rapidly restore growth under iron-limiting conditions. These adaptive events in experimental evolution depend on the siderophore biosynthetic function of S. fredii. Taken together, these findings suggest that the siderophore utilization ability of soybean rhizobia can be positively selected under iron-limiting conditions: by maintaining abundant xeno-siderophore receptors in acid-tolerant Bradyrhizobium and by the rapid adaptive evolution of utilization machinery for self-produced siderophores in alkaline-tolerant Sinorhizobium.
|
 Your new post is loading...
Your new post is loading...