 Your new post is loading...

|
Scooped by
?
Today, 1:08 AM
|
Galactomannan oligosaccharides (GMOS), composed of 2–10 mannose units linked with β-1, 4 glycosidic bond as the main chain and galactose linked with α-1, 6 glycosidic bond as the side chain, are crucial for probiotic food synthesis due to their ability to promote the growth and activity of beneficial intestinal microbiota, enhance the host immune system, and improve nutrient digestion. GMOS is usually obtained by hydrolyzing plants such as locust bean gum and guar gum with mannanase. β-mannanase ManA from Alkaliphilic Bacillus sp. N16-5 can hydrolyze β-1, 4 glycosidic bond of galactomannan. In this study, an immobilization system was employed utilizing polyhydroxyalkanoate (PHA) biopolymers, which naturally have an affinity mainly mediated by hydrophobic interaction for PhaP protein. Fusion protein combining ManA with PhaP from Aeromonas hydrophila, was subsequently immobilized on PHA support to form a multi-enzyme complex, facilitating the hydrolysis of locust bean gum to generate GMOS. This immobilized enzyme enhances enzyme stability and reusability, can be reused up to 32 times while maintaining ~ 80% of its activity, offering substantial cost savings through in-situ enzyme and product separation. Additionally, the different PHA forms were developed to hydrolyze locust bean gum to produce GMOS, such as nano PHA particles, PHA electrospun materials, while these preliminary investigations show promise, further research is needed to optimize their performance and practical application.

|
Scooped by
?
Today, 12:43 AM
|
Soybean-maize intercropping improves phosphate (Pi) acquisition in phosphorus (P) deficient soils through flavonoid-mediated plant-microbe interactions. Yet, the molecular mechanisms driving spatially heterogeneous root-microbe interactions mediated by secreted flavonoids remain unexplored. Using GmHAD1-2 suppression line (Ri) and wild-type (WT), we demonstrated that root-secreted flavonoids, particularly genistein, drive spatial differentiation of root allocation and rhizosphere microbial communities in intercropped soybean with maize, specifically under low-P conditions. Compared to WT, Ri reduced genistein secretion and restricted root allocation to the root non-interaction zone, thereby diminishing the intercropping advantage by less shoot biomass and P uptake. In all cropping systems, WT in intercropping recruited Bacillus in root non-interaction zones, while Pseudomonas in root interaction zones. Furthermore, inoculation experiments demonstrated their synergistic roles. Bacillus stimulated root elongation and enhanced transcription of auxin-responsive genes (i.e., GmPIN2b and GmYUC2a), whereas Pseudomonas elevated Pi availability in rhizosphere soils and upregulated Pi transporters (i.e., GmPHF1 and GmPT4). Taken together, spatial root allocation and heterogeneous microbial communities across root zones play a critical role in determining intercropping advantages, which is regulated by genistein exudation in soybean roots. Our study provides novel insights into root exudate-driven microbial zonation as a strategic adaptation to nutrient stress, with implications for optimising sustainable intercropping systems.

|
Scooped by
?
Today, 12:27 AM
|
The growing number of sequences and increasing proof that single references create reference bias have driven the development of pangenomes to represent the genomic diversity of species. To leverage this complex diversity information for biological insights, analysis and visualization support are needed to explore the variants in the context of metadata and phylogenies. We developed PanVA, an interactive visual analytics tool for exploring sequence variants in groups of homologous sequences in their biological context. PanVA is a web application that allows users to explore existing instances or create new ones to visualize their own data. PanVA source code is available on GitHub at https://github.com/PanBrowse/PanVA under the GPLv3 License. Documentation and and public demo instances showcasing examples can be accessed at https://panbrowse.github.io/PanVA/.

|
Scooped by
?
June 24, 11:57 PM
|
Arbuscular mycorrhizal fungi (AMF) are a key group of fungi closely associated with agricultural production within soil microbial communities. However, large-scale propagation of AMF inoculum faces various challenges, limiting our ability to obtain and utilize these inocula on a broad scale. To address this, we designed a monolayer mesh cultivation system employing a hydroponic approach for propagating arbuscular mycorrhizal fungi, specifically Rhizophagus intraradices. We conducted a comparative analysis of quality and inoculation efficiency between the water culture inoculum (w-Ri) and traditional soil-based inoculum (s-Ri). Our findings revealed the following. (i) The propagation cycle of w-Ri inoculum is 35 days and only 23% of the 150-day cycle required for s-Ri inoculum. (ii) The spore density, viability, and purity of w-Ri inoculum are 5.25 times, 1.09 times, and 1.26 times higher, respectively, than those of s-Ri inoculum. (iii) The w-Ri inoculants demonstrate effects on enhancing rice biomass, root morphology, and photosynthesis that are consistent with those of the s-Ri inoculants, while requiring only 10% of the application rate of the s-Ri inoculants. These results provide crucial theoretical references for establishing a pure and efficient arbuscular mycorrhizal fungus propagation system and its promotion and application.

|
Scooped by
?
June 24, 11:51 PM
|
There is a high demand for microbial pigments as a promising alternative for synthetic pigments, primarily for safety and economic reasons. This study aimed at the optimization of yellowish-orange pigment production by Exiguobacterium aurantiacum using agro-waste extracts as a growth substrate. Air samples were collected using the depositional method. Pure cultures of pigment producing bacteria were isolated by subsequent culturing on fresh nutrient agar medium. The potent isolate was identified using MALDI-TOF technique. Screening of culture conditions was done via Plackett-Burman design that highlighted culture agitation rate, initial medium pH, and yeast extract concentration as the most significant variables (p < 0.0001) in influencing pigment production with further optimization step using response surface methodology. Among the tested agro-waste decoctions, tomato waste extract was selected for fermentation due to higher optical density of the isolate when cultivated in it compared to the other agro-waste extracts. Under optimized conditions, 0.96 g/L of pigment was extracted from 4.73 g/L of culture biomass, representing a 1.6-fold increase compared to un-optimized conditions. Spectroscopic and chromatographic analyses confirmed the presence of various functional groups, with carotenoids identified as the primary compounds responsible for the yellowish-orange pigmentation. These findings demonstrate the feasibility of enhancing bacterial pigment production using agro-waste substrates, highlighting its potential for large-scale industrial applications.

|
Scooped by
?
June 24, 11:20 PM
|
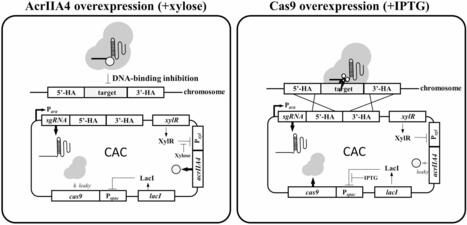
Wild-type Bacillus strains have significant industrial and medical value, but their effective utilization often requires strain improvement. The CRISPR/Cas9 system has become the primary tool for genome editing, allowing precise introduction of desired mutations at specific chromosomal locations. However, the practical application of CRISPR/Cas9 in most wild-type Bacillus strains remains challenging due to cellular toxicity resulting from Cas9/sgRNA activity. Therefore, controlling Cas9 toxicity is essential for the widespread application of the CRISPR/Cas9 system in wild-type Bacillus strains. We employed AcrIIA4, an anti-CRISPR protein that inhibits the Cas9/sgRNA ribonucleoprotein complex from interacting with DNA, to mitigate Cas9/sgRNA-mediated toxicity, thereby enabling CRISPR/Cas9-based genome editing in wild-type strains. The newly constructed CRISPR/anti-CRISPR (CAC) plasmids harbor both cas9 and acrIIA4 genes controlled by the Pspac and Pxyl promoters, respectively, along with the repressor genes lacI and xylR. This design allows precise control of Cas9 activity through inducers. Xylose, which induces AcrIIA4 expression, effectively alleviated Cas9/sgRNA-mediated toxicity during transformation. Under xylose induction, the CAC plasmid led to a remarkable 139-fold increase in the transformation efficiency of wild-type Bacillus subtilis compared to a plasmid lacking anti-CRISPR. Meanwhile, IPTG induction promoted Cas9 expression, facilitating efficient genome editing. Upon IPTG induction, the genome editing efficiency in wild-type B. subtilis increased from 0 to 95.8% in transformants carrying the CAC plasmid. Importantly, our findings extend beyond B. subtilis, revealing that the anti-CRISPR protein dramatically enhanced transformation and genome editing efficiencies in Bacillus pumilus. Moreover, we demonstrated that the CAC system successfully enabled the generation of spo0A mutants in Bacillus mojavensis, Bacillus tequilensis, and Paenibacillus polymyxa. Conclusions In this study, we developed a CAC system that utilizes the anti-CRISPR protein AcrIIA4 to reduce Cas9/sgRNA-mediated toxicity in Bacillus strains. This system enables precise control of AcrIIA4 and Cas9 expression through inducers, significantly enhancing the efficiency of transformation and genome editing in wild-type Bacillus strains. Therefore, the CAC system stands as a powerful tool to facilitate genome editing in diverse wild-type Bacillus species.

|
Scooped by
?
June 24, 10:49 PM
|
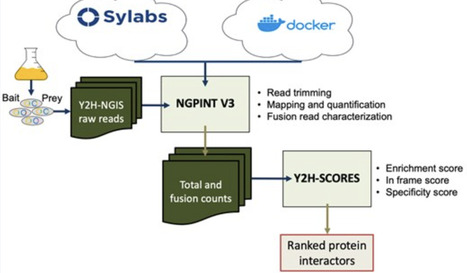
Batch yeast two-hybrid (Y2H) assays, leveraged with next-generation sequencing (NGS), have afforded successful innovations for the analysis of protein-protein interactions (PPIs). NGPINT is a Conda-based software designed to process the millions of raw sequencing reads resulting from yeast two hybrid-next generation interaction screens (Y2H-NGIS). Over time, increasing compatibility and dependency issues have prevented clean NGPINT installation and operation. A system-wide update was essential to continue effective use with its companion software, Y2H-SCORES. We present NGPINT V3, a containerized implementation built with both Singularity and Docker, allowing accessibility across virtually any operating system and computing environment. This update includes streamlined dependencies and container images hosted on Sylabs (https://cloud.sylabs.io/library/schuyler/ngpint/ngpint) and Dockerhub (https://hub.docker.com/r/schuylerds/ngpint), facilitating easier adoption and integration into high-throughput and cloud-computing workflows. Full instructions and software can be also found in the GitHub repository https://github.com/Wiselab2/NGPINT_V3 and Zenodo https://doi.org/10.5281/zenodo.15256036.

|
Scooped by
?
June 24, 10:59 AM
|
Concerns over the depletion of fossil resources prompt us to consider the development of green and energy-saving resources. One alternative to fossil fuels is the microbial biosynthesis of chemicals using renewable carbon sources through metabolic engineering. In this review, we provide a broad and high-level overview of various research efforts to address the challenges of utilizing sustainable carbon sources, including glucose, lignocellulose, and one‑carbon (C1) compounds, through synthetic biology. We emphasize these endeavors can accelerate the development of microbial industrial application. Additionally, we discuss the prospects of using multi-omics sequencing, machine learning, and artificial intelligence to guide strain engineering for improving the utilization of sustainable carbon sources and prospects for reducing their costs.

|
Scooped by
?
June 24, 10:45 AM
|
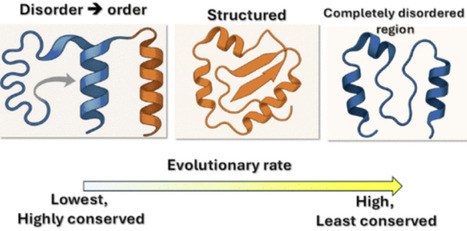
Protein structure is a major determinant of protein evolution at the residue level. It is especially challenging to elucidate the relationship between structure and evolution for proteins that adopt several different conformations due to their function. Previously, we showed that proteins that switch between different native states are on average under stronger selective pressure than proteins with a single native state. Here, we analyzed evolutionary rates at the residue level for proteins that switch between disordered and ordered states in the yeast proteome. We show that while proteins with completely disordered regions generally evolve more rapidly than structured proteins, proteins with disorder–order switching regions evolve significantly more slowly on average. Surprisingly, proteins with disorder–order switching regions on average evolve even more slowly than conformational switches, which are known to be highly conserved. The elevated selective pressure on proteins with disorder–order switching regions is exerted on the entire protein, such that even the ordered residues in these proteins are on average more conserved than the ordered residues in structured proteins. While disordered residues generally evolve more rapidly than ordered residues within a protein, disordered regions that can switch into ordered states are highly conserved compared with other disordered residues. Overall, our results suggest that the necessity to encode and maintain coupled folding and binding imposes a unique and strong selective pressure on the entire protein.

|
Scooped by
?
June 24, 10:25 AM
|
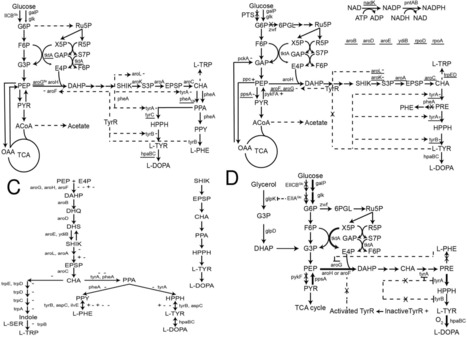
l-DOPA (3,4-dihydroxyphenyl-l-alanine) has been the primary medication for treating Parkinson's disease (PD), a degenerative brain disorder related to dopamine depletion, for the past six decades. As a result, biotechnological approaches utilizing metabolic engineering in microorganisms or enzymatic processes have been extensively explored as promising alternatives for l-DOPA production. These methods not only enhance conversion efficiency and enantioselectivity but are also cost-effective and environmentally sustainable. Metabolic engineering strategies have been employed to engineer Escherichia coli strains capable of accumulating l-DOPA from glucose by regulating carbon metabolism pathways. Additionally, microbial systems expressing tyrosinase, p-hydroxyphenylacetate 3-hydroxylase (PHAH), or tyrosine phenol-lyase (TPL) have been utilized for l-DOPA biosynthesis. In this review, we summarize current advancements in l-DOPA biosynthesis and biotechnological production strategies, providing a comparative analysis of their advantages and limitations. Moreover, we discuss the promise of biotech-driven l-DOPA production, emphasizing its industrial applications and large-scale production feasibility.

|
Scooped by
?
June 24, 10:20 AM
|
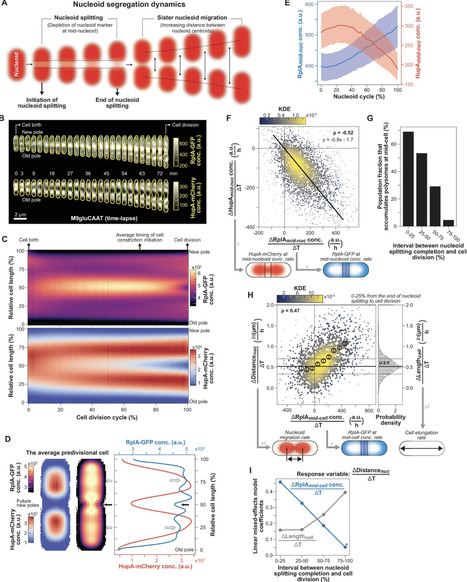
Chromosome segregation is essential for cellular proliferation. Unlike eukaryotes, bacteria lack cytoskeleton-based machinery to segregate their chromosomal DNA (nucleoid). The bacterial ParABS system segregates the duplicated chromosomal regions near the origin of replication. However, this function does not explain how bacterial cells partition the rest (bulk) of the chromosomal material. Furthermore, some bacteria, including Escherichia coli, lack a ParABS system. Yet, E. coli faithfully segregates nucleoids across various growth rates. Here, we provide theoretical and experimental evidence that polysome production during chromosomal gene expression helps compact, split, segregate, and position nucleoids in E. coli through nonequilibrium dynamics that depend on polysome synthesis, degradation (through mRNA decay), and exclusion from the DNA meshwork. These dynamics inherently couple chromosome segregation to biomass growth across nutritional conditions. Halting chromosomal gene expression and thus polysome production immediately stops sister nucleoid migration, while ensuing polysome depletion gradually reverses nucleoid segregation. Redirecting gene expression away from the chromosome and toward plasmids causes ectopic polysome accumulations that are sufficient to drive aberrant nucleoid dynamics. Cell width enlargement experiments suggest that limiting the exchange of polysomes across DNA-free regions ensures nucleoid segregation along the cell length. Our findings suggest a self-organizing mechanism for coupling nucleoid compaction and segregation to cell growth without the apparent requirement of regulatory molecules.

|
Scooped by
?
June 24, 10:03 AM
|
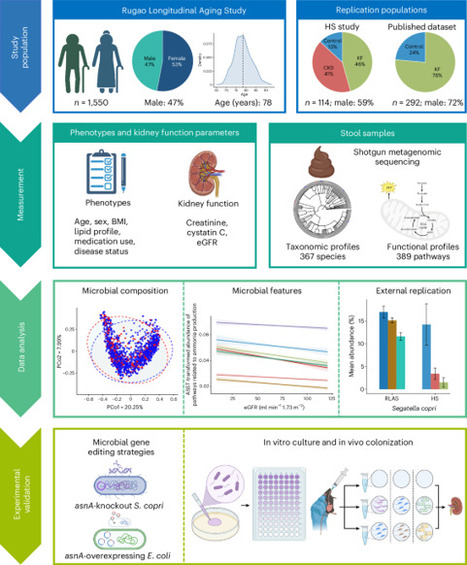
Alterations in gut microbiota have been linked to chronic kidney disease (CKD), but large-scale studies and mechanistic insights are limited. Here we analyzed gut metagenome data from 1,550 older individuals (aged 65–93 years) with comprehensive kidney function measurements. Segatella copri was positively associated with kidney function through microbial ammonia metabolism-related pathways and the asnA gene, which encodes an ammonia-assimilating enzyme. These associations were replicated in two external studies. In mice, ammonia supplementation increased serum levels of creatinine and blood urea nitrogen, accelerating CKD progression. In vitro cultures of S. copri or asnA-overexpressing Escherichia coli reduced ammonia concentrations, which was markedly attenuated in asnA-knockout S. copri. Gavage of either S. copri or asnA-overexpressing E. coli, but not asnA-knockout S. copri, mitigated ammonia-induced CKD progression in mice. These findings highlight the role of gut microbial ammonia metabolism in CKD pathogenesis and underscore the therapeutic potential of microbial-based interventions. Using human data and mice models, it was revealed that assimilation of ammonia by a gut microbe prevents its harmful accumulation that can impair kidney function.

|
Scooped by
?
June 24, 9:51 AM
|
RNA sequencing (RNA-seq) has become an essential technology for assessing gene expression profiles in biomedical research. However, the coding complexity of RNA-seq data analysis remains a significant barrier for students and researchers without extensive bioinformatics expertise. We present the Educational RNA-Seq Analysis tool (ERSAtool), a comprehensive R/Shiny interface that provides an intuitive graphical visualization of the complete RNA-seq analysis workflow. The application is built on established Bioconductor packages and upholds high standards in analyses while significantly reducing the technical expertise required to conduct sophisticated transcriptomic analyses. ERSAtool supports various input formats, such as raw count matrices and STAR alignment outputs. It generates sample information metadata through direct integration with the Gene Expression Omnibus (GEO) provided by the National Center for Biotechnology Information (NCBI). The application guides users through normalization, data visualization, differential expression analysis, and functional interpretation using Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA). All results can be compiled into comprehensive, downloadable reports that enhance reproducibility and knowledge sharing. The design includes targeted features that facilitate educational use, making it especially useful for teaching transcriptomics in undergraduate to graduate-level bioinformatics courses. By connecting command-line bioinformatics tools with accessible graphical interfaces, ERSAtool improves accessibility to advanced transcriptomic analysis capabilities, potentially accelerating discoveries across various biological fields. https://github.com/SuzukiLabTAMU/ERSAtool
|

|
Scooped by
?
Today, 1:00 AM
|
Prokaryotic defense-associated reverse transcriptases (DRTs) were recently identified with antiviral functions; however, their functional mechanisms remain largely unexplored. Here we show that DRT9 forms a hexameric complex with its upstream noncoding RNA (ncRNA) to mediate antiphage defense by inducing cell growth arrest through abortive infection. Upon phage infection, the phage-encoded ribonucleotide reductase NrdAB complex increases intracellular deoxyadenosine triphosphate levels, activating DRT9 to synthesize long, polyadenylate [poly(A)]–rich single-stranded complementary DNA (cDNA), which likely sequesters the essential phage single-stranded DNA binding (SSB) protein and disrupts phage propagation. We further determined the cryo–electron microscopy structure of the DRT9-ncRNA hexamer complex, providing mechanistic insights into its cDNA synthesis. These findings highlight the diversity of RT-based antiviral defense mechanisms, expand our understanding of RT biological functions, and provide a structural basis for developing DRT9-based biotechnological tools.

|
Scooped by
?
Today, 12:30 AM
|
Protein-peptide interactions mediate many biological processes, and access to accurate structural models, through experimental determination or reliable computational prediction, is essential for understanding protein function and designing novel protein-protein interactions. AlphaFold2-Multimer (AF2-Multimer), AlphaFold3 (AF3), and related models such as Boltz-1 and Chai-1 are state-of-the-art protein structure predictors that successfully predict protein-peptide complex structures. Using a dataset of experimentally resolved protein-peptide structures, we analyzed the performance of these four structure prediction models to understand how they work. We found evidence of bias for previously seen structures, suggesting that models may struggle to generalize to novel target proteins or binding sites. We probed how models use the protein and peptide multiple sequence alignments (MSAs), which are often shallow or of poor quality for peptide sequences. We found weak evidence that models use coevolutionary information from paired MSAs and found that both the target and peptide unpaired MSAs contribute to performance. Our work highlights the promise of deep learning for peptide docking and the importance of diverse representation of interface geometries in the training data for optimal prediction performance.

|
Scooped by
?
Today, 12:19 AM
|
Biofilms are groups of microbes that live together in dense communities, often attached to a surface. They play an outsized role in all aspects of microbial life, from chronic infections to biofouling to dental decay. In recent decades, appreciation for the diversity of roles that biofilms play in the environment has grown. Yet, most bacterial studies still rely upon approaches developed in the 19th century and center on planktonic populations alone. Here we present a chemostat-based experimental platform to investigate not only biofilms themselves, but how they interact with their surrounding environments. Our results show that biofilms grow to larger sizes in chemostats as opposed to flasks. In addition, we show that biofilms may be a consistent source of migrants into planktonic populations. We also show that secondary biofilms rapidly develop, although these may be more susceptible to environmental conditions. Taken together, our data suggest that chemostats may be a flexible and insightful platform for the study of biofilms in vitro.

|
Scooped by
?
June 24, 11:53 PM
|
CRISPR-Cas9 is a powerful tool for gene editing and regulation, facilitating the analysis of gene function. In this study, we developed a robust CRISPR interference (CRISPRi) system to precisely modulate gene expression in the bacterium Rhizobium etli, the nitrogen-fixing symbiont of the common bean. The system is based on two compatible plasmids (pBBR1MCS2-dCas9 and pRhigRNA containing specific guide RNAs). Introduction of both plasmids in R. etli led to significant repression of four target genes [DsRedexpress, recA, thiC (on the thiCOSGE operon) and rdsA] depending on the guide RNA used. By employing different guide RNAs at various target sites, we obtained up to 90% gene repression. Importantly, neither significant secondary effects on growth nor toxicity were observed upon expression of dCas9, either alone or in co-expression with the guide RNAs. This system can be utilized for further investigations on the function of essential genes in R. etli, or it can be integrated with other gene expression elements or gene editing tools, such as base editors for advanced genome engineering in Rhizobiales.

|
Scooped by
?
June 24, 11:22 PM
|
Integrons are genetic elements that facilitate gene acquisition. They have been extensively studied in clinical bacteria, but their evolutionary role in phytopathogens remains underexplored. Here, we analysed complete genomes of Xanthomonas species to investigate the origin, distribution, and functional dynamics of integrons in this genus. We found that 93% of genomes harbored integrons. The integron-integrase gene intI was predominantly located downstream of ilvD, indicating an ancestral acquisition of integrons, predating diversification within the genus. Phylogenetic analyses support vertical inheritance of intI, with the exception of rare horizontal gene transfer events, notably in X. arboricola. Despite their widespread presence, full-length intI genes and active integron platforms are only retained in some species, especially X. campestris, which shows high integron gene cassette variability and functional integron activity. In contrast, species such as X. cissicola and X. phaseoli exhibit widespread inI inactivation, likely occurring early in their divergence, leading to more stable cassette arrays and conserved integron-associated phenotypes. The number and diversity of genes within cassette arrays varied significantly by species and, to a lesser extent, by the ecological context of plant host cultivation. While most cassettes encoded proteins without a known function, those with annotated roles were associated with stress response mechanism, competitive exclusion, and plant-associated functions. Together, our findings demonstrate that integrons in Xanthomonas likely originated from a single ancient acquisition event, preceding genus-wide speciation, and have co-evolved with Xanthomonas pathovars as they adapted to distinct plant hosts.

|
Scooped by
?
June 24, 10:56 PM
|
Establishing efficient microbial cell factories for the production of functional nutraceuticals, pharmaceuticals, biofuels, and chemical products requires precise regulation to adapt key enzymes and pathway modules. Dynamic regulatory strategies are a promising and effective approach to achieve balanced cell growth and metabolite production. Dynamic regulatory tools, as the executors of regulatory strategies, usually require rationally designed modification strategies to provide libraries of tools with reliable quality. Here, typical dynamic regulatory tools at the DNA level (transcriptional level), the RNA level (post-transcriptional and translational level), and the protein level (post-translational) are presented. The regulatory mechanisms and design modification strategies of each tool are highlighted. Subsequently, strategies for applying regulatory tools to construct dynamic regulatory networks of metabolic pathways are summarized. Finally, the limitations of current dynamic regulatory tools are discussed and future trends are outlooked.

|
Scooped by
?
June 24, 11:02 AM
|
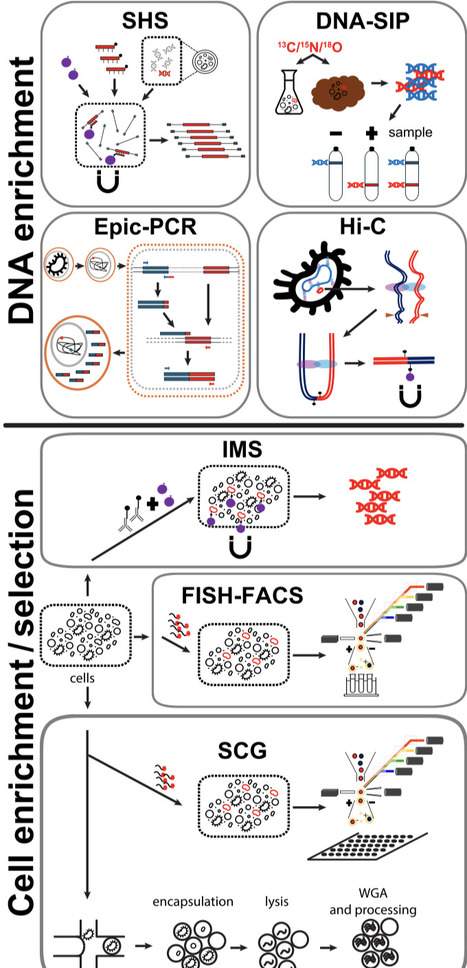
The rapidly developing field of targeted analysis of metagenomes focuses on retrieving information about specific genes and/or genome(s) from environmental DNA. The traditional shotgun sequencing methods over-emphasize dominant microorganisms and often fail to confidently assign the entirety of the analyzed genetic material to specific species, genomovars, or strains. The ultimate goal of the targeted methods is to overcome this limitation of throughput and precision of current shotgun metagenomics when analysing complex microbial communities in the quest of refined information. Here, we discuss recent technological advances that are designed to focus the analytical power of diagnostic tools like sequencing, towards phylogenetically or functionally distinct and rare microbial groups and enhance e.g. the confidence in the assignment of genetic elements to their respective owning organisms. We specifically showcase the capabilities of these technological advances for targeted analysis of metagenomes, identify suitable related applications, discuss methodological limitations, and propose solutions for addressing these limitations. This review aspires to inspire highly relevant experimental designs in the future that will unlock unknown and important aspects of microbial ecology, and the yet-uncultivated microbial majority.

|
Scooped by
?
June 24, 10:52 AM
|
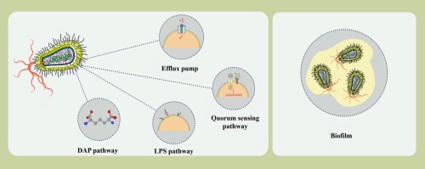
Pseudomonas aeruginosa, an opportunistic pathogen known for its adaptability, has become a critical health concern due to its inherent resistance to multiple antibiotic classes and its rapid acquisition of new resistance mechanisms. The rise of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains has further compounded the global burden of P. aeruginosa infections. Traditional antibiotic discovery efforts, which focus on essential bacterial processes such as cell wall synthesis, protein production, and DNA replication, have been unable to keep pace with the pathogen's evolving resistance strategies. Recent advancements in omics technologies have provided deeper insights into the complex biology of P. aeruginosa, including bacterial communication networks like quorum sensing and interactions between the host and pathogen that are crucial for the pathogen's survival and virulence. These insights pave the way for identifying novel therapeutic targets, such as unexplored metabolic pathways and virulence mechanisms, which could offer more effective strategies for combating resistant P. aeruginosa strains. In this review, we critically assess the limitations of conventional approaches and emphasize the potential of targeting these alternative pathways to address the growing challenge of antibiotic resistance. By exploring innovative strategies that transcend traditional methods, this review underscores the importance of pursuing novel therapeutic avenues that could lead to the development of more effective antibiotics against P. aeruginosa and similar resistant pathogens.

|
Scooped by
?
June 24, 10:33 AM
|
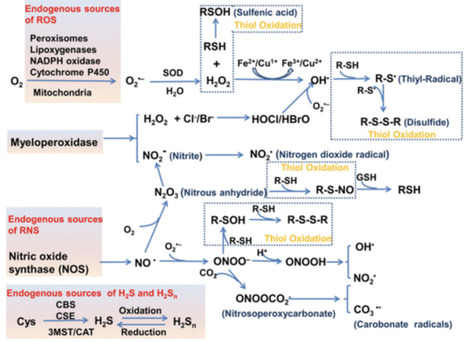
Cellular redox homeostasis governs numerous essential biological processes and presents broad implications in human health and diseases. Cells maintain the redox homeostasis by expression of some types of oxidants, reductants, and redox-active metals, while processing elaborate mechanisms to regulate their internal redox status. These biomolecules can serve as biomarkers to identify the alterations in redox status. The cellular redox dysregulation indicates a crucial pathogenic mechanism in conditions such as inflammatory diseases, cancer, and neurodegenerative disorders. Consequently, exploring the self-regulatory mechanisms of cellular redox homeostasis and in situ imaging of redox changes have emerged as key research focuses. In this review, we focus on the biomolecules involved in redox maintenance and highlight their detailed regulatory mechanisms. Moreover, we systematically review recent progress in fluorescent probes responsive to biomarkers of redox changes, primarily over the past five years, including their design principles, reaction mechanisms, and bioimaging applications in redox-related diseases. This review concludes with a discussion of the challenges and prospects for redox regulation and imaging in the therapeutics and diagnostics of human diseases. We hope it can serve as a valuable resource for those interested in this rapidly expanding research field.

|
Scooped by
?
June 24, 10:22 AM
|
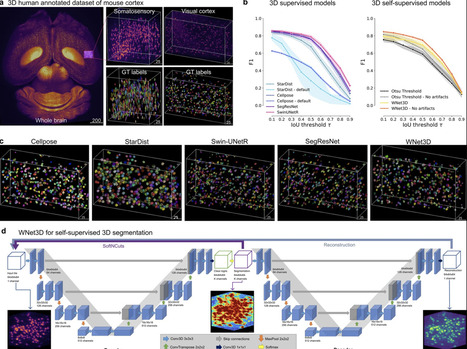
Understanding the complex three-dimensional structure of cells is crucial across many disciplines in biology and especially in neuroscience. Here, we introduce a set of models including a 3D transformer (SwinUNetR) and a novel 3D self-supervised learning method (WNet3D) designed to address the inherent complexity of generating 3D ground truth data and quantifying nuclei in 3D volumes. We developed a Python package called CellSeg3D that provides access to these models in Jupyter Notebooks and in a napari GUI plugin. Recognizing the scarcity of high-quality 3D ground truth data, we created a fully human-annotated mesoSPIM dataset to advance evaluation and benchmarking in the field. To assess model performance, we benchmarked our approach across four diverse datasets: the newly developed mesoSPIM dataset, a 3D platynereis-ISH-Nuclei confocal dataset, a separate 3D Platynereis-Nuclei light-sheet dataset, and a challenging and densely packed Mouse-Skull-Nuclei confocal dataset. We demonstrate that our self-supervised model, WNet3D – trained without any ground truth labels – achieves performance on par with state-of-the-art supervised methods, paving the way for broader applications in label-scarce biological contexts.

|
Scooped by
?
June 24, 10:06 AM
|
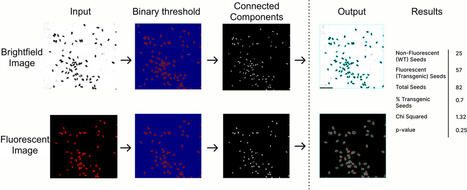
Transgenic plants are essential for both basic and applied plant biology. Recently, fluorescent and colorimetric markers were developed to enable nondestructive identification of transformed seeds and accelerate the generation of transgenic plant lines. Yet, transformation often results in the integration of multiple copies of transgenes in the plant genome. Multiple transgene copies can lead to transgene silencing and complicate the analysis of transgenic plants by requiring researcher to track multiple T-DNA loci in future generations. Thus, to simplify analysis of transgenic lines, plant researchers typically screen transformed plants for lines where the T-DNA inserted in a single locus — an analysis that involves laborious manual counting of fluorescent and non-fluorescent seeds for screenable markers. To expedite T-DNA segregation analysis, we developed SeedSeg, an image analysis tool that uses a segmentation algorithm to count the number of transformed and wild-type seeds in an image. SeedSeg runs a chi-squared test to determine the number of T-DNA loci. Parameters can be adjusted to optimize for different brightness intensities and seed sizes. By automating the seed counting process, SeedSeg reduces the manual labor associated with identifying transgenic lines containing a single T-DNA locus. SeedSeg is adaptable to different seed sizes and visual transgene markers, making it a versatile tool for accelerating plant research.

|
Scooped by
?
June 24, 9:55 AM
|
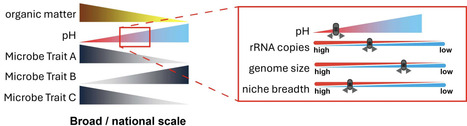
Understanding the relationships between bacteria, their ecological and genomic traits, and their environment, is important to elucidate microbial community dynamics and their roles in ecosystem functioning. Here, we examined the relationships between soil properties and bacterial traits within highly managed agricultural soil systems subjected to arable crop rotations or management as permanent grass. We assessed the bacterial communities using metabarcoding and assigned each amplicon trait scores for rRNA copy number, genome size, and GC content, which are classically associated with potential growth rates and specialisation. We also calculated the niche breadth trait of each amplicon as a measure of social ubiquity within the examined samples. Within this soil system, we demonstrated that pH was the primary driver of bacterial traits. The weighted mean trait scores of the samples revealed that bacterial communities associated with soils at lower pH (<7) tended to have larger genomes (potential plasticity), have more rRNA (higher growth rate potential), and are more ubiquitous (have less niche specialisation) than the bacterial communities from higher pH soils. Our findings highlight not only the association between pH and bacterial community composition but also the importance of pH in driving community functionality by directly influencing genomic and niche traits.
|
 Your new post is loading...
Your new post is loading...



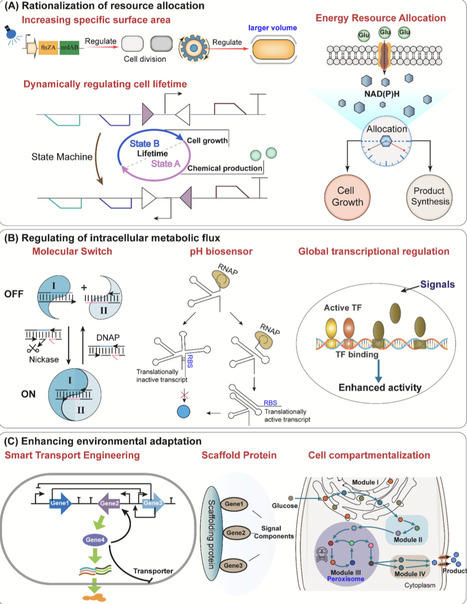

















1str, cell size, by screening genes that shorten the C and D phases of cell division, blue light activation and near-infrared light activation systems were constructed to regulate cell division to increase the specific surface area of E. coli, thereby increasing the production of acetoin to 67.2 g/L. Similarly, cell division was reversibly spatiotemporal regulated to increase the cell volume of E. coli, thereby increasing the production of poly (lactate-co-3-hydroxybutyric acid) to 14.3 g/L. On the other hand, it can also improve the production performance of cell factories by delaying cell aging. Key targets of E. coli sequential lifetime (ubiG, rssB, and rpoS) and replication lifetime (csrA) were precisely regulated by logic gate state machines to achieve 29.8 g/L yield of butyric acid and asymmetric distribution of poly (lacto-co-3-hydroxybutyric acid) in vivo