Your new post is loading...

|
Scooped by
?
Today, 1:36 PM
|
Synthetic biology can endow microorganisms with novel capabilities. Su et al. developed an engineered strain of Vibrio natriegens that efficiently degrades complex organic pollutants in high-salinity environments. This technology offers a new biotechnological solution to urgent global environmental challenges, such as petrochemical wastewater discharges and marine oil spills.

|
Scooped by
?
Today, 1:28 PM
|
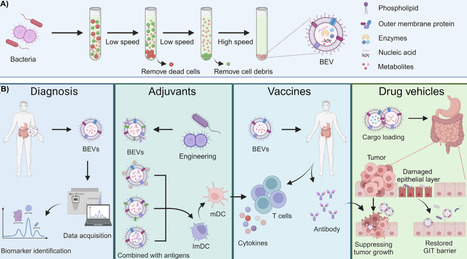
Bacterial extracellular vesicles (BEVs) are nanoscale spherical particles with lipid bilayer membranes containing diverse functional components from their parent bacteria. They exert pivotal effects on bacteria–bacteria and host–bacteria communication. Analyzing the dynamic changes in the production and composition of BEVs provides insights into the relationship between gut microbiota and host health, offering valuable perspectives for diagnosing various gastrointestinal diseases. Furthermore, BEVs can be employed as natural medications and drug delivery vehicles for inflammation management, cancer treatment, and vaccine development. This review summarizes the structural composition, generation mechanism, and biomedical applications of BEVs, emphasizing recent advances in immune regulation, gut microbiota modulation, and the clinical translation challenges associated with gastrointestinal diseases.

|
Scooped by
?
Today, 1:25 PM
|
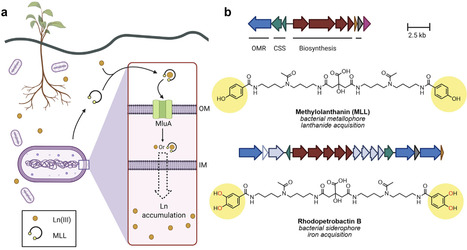
Metallophores are metal-chelating secondary metabolites that bacteria, fungi, and other organisms produce to acquire essential metal nutrients from their environment. This review highlights some recent discoveries in the field: the role of yersiniabactin in quorum sensing, the capacity of aspergillomarasmine A to inhibit bacterial Ni uptake, and the identification of methylolanthanin, a novel metallophore that enables bacterial lanthanide acquisition.

|
Scooped by
?
Today, 1:07 PM
|
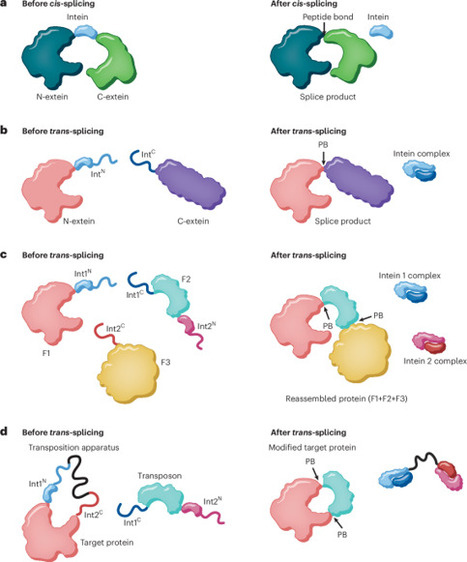
Transposition is normally considered a DNA-based process. Now, scientists have devised a means to carry out a similar transformation — this time using proteins. The approach enables protein semisynthesis and site-specific modification of folded substrates in vitro and in live cells, opening up new ways to probe and manipulate biological systems.

|
Scooped by
?
Today, 1:27 AM
|
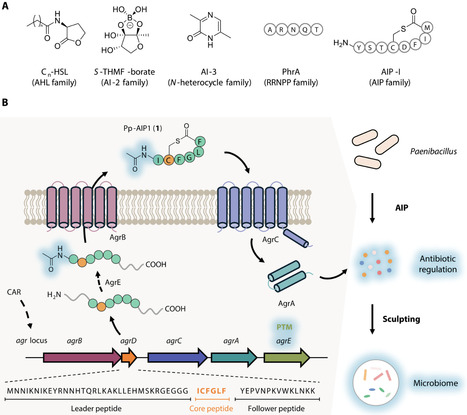
Microbes use signaling molecules to regulate multiple physiological processes and mediate chemical interactions. Decoding these chemical languages is instrumental in comprehending microbial regulatory mechanisms within complex microbiota. Here, we discover previously unidentified autoinducing peptides (AIPs) derived from the plant probiotic bacterium Paenibacillus polymyxa, identified as Pp-AIPs. Omics analyses coupled with genetic manipulations revealed that Pp-AIP1 could effectively modulate the production of multiple antimicrobial secondary metabolites at nanomolar concentration, expanding known AIP functions. Furthermore, through inoculating P. polymyxa in the natural rhizosphere microbiome and analyzing its antagonistic interactions against root microbes, we suggest that Pp-AIPs may influence the microbial community composition through modulating the antimicrobial spectrum. Global analysis of biosynthetic gene clusters (BGCs) reveal widespread co-occurrence of uncharacterized AIPs with secondary metabolite BGCs. This study underscores the unreported roles of AIPs in antibiotic regulation and the microbiome interactions, advancing knowledge of quorum-sensing mechanisms in microbial ecosystems.

|
Scooped by
?
Today, 12:59 AM
|
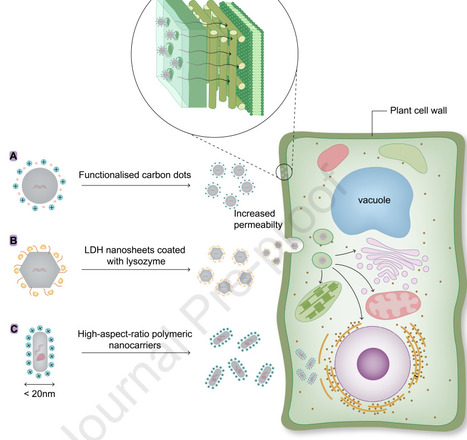
Nano-carriers are revolutionizing plant genetic engineering by facilitating the direct and efficient delivery of nucleic acids and proteins into plant cells. By precisely controlling and tailoring their properties, these carriers can effectively penetrate the plant cell wall, delivering bioactive molecules overcoming the constraints imposed by plant genotype specificity. This advancement provides a powerful, flexible, and broadly applicable strategy for accelerating crop health and resilience. Conventional plant transformation methods, such as Agrobacterium-mediated gene transfer and biolistic delivery, are limited by species-specific constraints, lengthy regeneration processes, and increased risks of genomic instability. Nanomaterial-based delivery systems offer a flexible alternative but face the significant challenge of penetrating the complex, multilayered plant cell wall. Recent research highlights three effective strategies: optimizing particle size, adjusting surface charge, and using cell wall-degrading enzymes. Together, these approaches greatly enhance nanocarrier uptake, enabling delivery of diverse bioactive agents like nucleic acids, proteins and gene-editing tools making nanotechnology a promising tool for crop improvement.

|
Scooped by
?
Today, 12:39 AM
|
Bacterial genomes exhibit significant variation in gene content and sequence identity. Pangenome analyses explore this diversity by classifying genes into core and accessory clusters of orthologous groups (COGs). However, strict sequence identity cutoffs can misclassify divergent alleles as different genes, inflating accessory gene counts. CLARC (Connected Linkage and Alignment Redefinition of COGs) (https://github.com/IndraGonz/CLARC) improves pangenome analyses by condensing accessory COGs using functional annotation and linkage information. Through this approach, orthologous groups are consolidated into more practical units of selection. Analyzing 8000+ Streptococcus pneumoniae genomes, CLARC reduced accessory gene estimates by >30% and improved evolutionary predictions based on accessory gene frequencies. CLARC is effective across different bacterial species, making it a broadly applicable tool for comparative genomics. By refining COG definitions, CLARC offers critical insights into bacterial evolution, aiding genetic studies across diverse populations.

|
Scooped by
?
Today, 12:22 AM
|
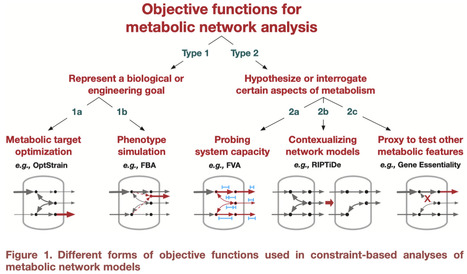
The “objective function” is a core concept in metabolic network modeling. Its use has enabled the analysis of large data to drive deeper understanding of cellular metabolism. This commentary reframes how the objective function is discussed to enhance its value and clarify misunderstandings in metabolic network modeling.

|
Scooped by
?
Today, 12:14 AM
|
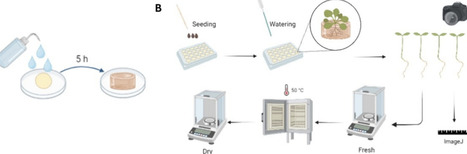
Hydrogels are emerging as sustainable alternatives to petroleum-based foams and pots in horticulture due to their high porosity, enhancing water and nutrient retention. This study develops κ-carrageenan (Carr)-based hydrogels incorporating hydrolyzed (HSW) and nonhydrolyzed red seaweed (WSW) (0–50 wt %) to introduce biostimulant properties for soilless cultivation. Hydrogels were analyzed for swelling in different media, solubility, microstructure (SEM), physicochemical interactions (FTIR, XRD), and mechanical properties, revealing high porosity, superabsorbent behavior (swelling up to 6000%), and reduced compression modulus with increasing seaweed content. Their biostimulant effect was assessed on Arabidopsis thaliana, with 20 wt % HSW hydrogels promoting enhanced root and shoot growth. Additionally, the hydrogels inhibited Fusarium solani, demonstrating antifungal properties. These results highlight the potential of Carr-seaweed hydrogels as multifunctional substrates for sustainable plant cultivation.

|
Scooped by
?
June 21, 4:59 PM
|
Pseudomonas putida is an attractive synthetic biology platform organism for chemical synthesis from low-grade feedstocks due to its high tolerance to chemical solvents and lignin-derived small molecules that are often inhibitory to other biotechnologically relevant microorganisms. However, there are few molecular tools available for engineering P. putida and other gram-negative bacteria to secrete non-native enzymes for extracellular feedstock depolymerisation. In this study P. putida was transformed to secrete cellulase enzymes and evaluated for growth on polymeric or oligomeric cellulose substrates. Active exo- and endocellulase enzymes were secreted into the culture supernatant, and a preferred set of twin-arginine translocase secretion signal peptides were identified. Extracellular cellulase activity was sufficient to support growth of P. putida using cellotriose or cellotetraose as the sole source of carbon and energy. This work supports progress towards consolidated bioprocessing of cellulosic materials using P. putida, and advances the state of engineered protein secretion in gram negative bacteria.

|
Scooped by
?
June 21, 4:33 PM
|
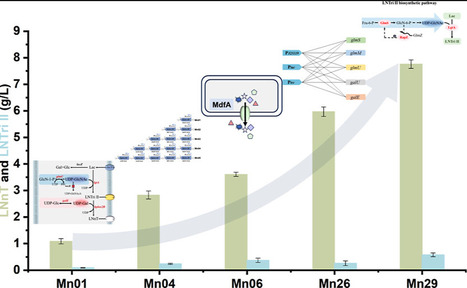
Lacto-N-neotetraose (LNnT) is a key human milk oligosaccharide (HMO) with important prebiotic functions, supporting the growth of beneficial gut microbiota and contributing to infant health. Constructing plasmid-free strains via metabolic engineering for LNnT biosynthesis represents a feasible strategy for efficient industrial-scale production. This study integrates various strategies to construct plasmid-free strains capable of efficiently producing LNnT. Building on the previously developed Escherichia coli MG1655 strain for lacto-N-triose II (LNTri II) production, Hplex2B (encoding β1,4-galactosyltransferase) was incorporated, and its copy number was optimized to construct a complete and efficient biosynthetic pathway. By integrating an extra copy of the multidrug efflux pump gene mdfA, the strain tolerance was improved, resulting in a higher yield of LNnT. The integration of expression cassettes for key genes in the glycosyl donor synthesis pathway (including glmS, glmM, glmU, galU, and galE) with varying promoter strengths optimized the supply and balance. The subsequent removal of a key feedback inhibition circuit directed the reaction toward the synthesis of LNnT. The final strain, after successive optimization, produced LNnT at 7.76 g/L in shake flask culture and 34.24 g/L in a 5 L bioreactor, with precursor LNTri II concentrations of 0.58 g/L and 1.61 g/L, respectively.

|
Scooped by
?
June 21, 4:10 PM
|
Biofilm formation allows pathogenic nontuberculous mycobacteria (NTM) to adhere to household plumbing systems and has been observed in vivo during human infection. Glucose drives NTM aggregation in vitro, and ammonium inhibits it, but the regulatory systems controlling this early step in biofilm formation are not understood. Here, we show that a variety of carbon and nitrogen sources have similar impacts on aggregation in the model NTM Mycobacterium smegmatis as glucose and ammonium, suggesting that the response to these nutrients is general and likely sensed through downstream, integrated signals. Next, we performed a transposon screen in M. smegmatis to uncover these putative regulatory nodes. Our screen revealed that mutating specific genes in the purine and pyrimidine biosynthesis pathways caused an aggregation defect, but supplementing with adenosine and guanosine had no impact on aggregation either in a purF mutant or WT. Realizing that the only genes we hit in purine or pyrimidine biosynthesis were those that utilized glutamine as a nitrogen donor, we pivoted to the hypothesis that intracellular glutamine could be a nitrogen-responsive node affecting aggregation. We tested this hypothesis in defined M63 medium using targeted mass spectrometry. Indeed, intracellular glutamine increased with nitrogen availability and correlated with planktonic growth. Furthermore, a garA mutant, which has an artificially expanded glutamine pool in growth phase, grew solely as planktonic cells even without nitrogen supplementation. Altogether these results establish that intracellular glutamine controls M. smegmatis aggregation, and they introduce flux-dependent sensors as key components of the NTM biofilm regulatory system.

|
Scooped by
?
June 21, 3:54 PM
|
FastGA finds alignments between two genome sequences more than an order of magnitude faster than previous methods that have comparable sensitivity. Its speed is due to (a) a carefully engineered architecture involving only cache-coherent MSD radix sorts and merges, (b) a novel algorithm for finding adaptive seed hits in a linear merge of sorted k-mer tables, and (c) a variant of the Myers adaptive wave algorithm to find alignments around a chain of seed hits that detects alignments with up to 25-30% variation. It further does not require pre-masking of repetitive sequence, and stores millions of alignments in a fraction of the space of a conventional CIGAR-string using a trace-point encoding that is further compressed by the ONEcode data system introduced here. As an example, two bat genomes of size 2.2Gbp and 2.5Gbp can be compared in a little over 2 minutes using 8 threads on an Apple M4 Max laptop using 5.7GB of memory and producing 1.05 million alignments totaling 1.63Gbp of aligned sequence that cover about 60% of each genome. The output ALN-formated file occupies 66MB. This file can be converted to a PAF file with CIGAR strings in 6 seconds, where the PAF representation is a significantly larger 1.03GB file. FastGA is available: http://www.github.com/thegenemyers/FASTGA, along with utilities for viewing inputs, intermediate files, and outputs and transforming outputs into other common formats. Specifically, FastGA can, in addition to its highly efficient ONEcode representation, output PSL-formatted alignments, or PAF-formatted alignments with or without CIGAR strings explicitly encoding the alignments. There is also a utility to chain FastGA's alignments and display them in a dot-plot like view in Postscript files, and an interactive viewer is in development.
|

|
Scooped by
?
Today, 1:31 PM
|
Cyanidiophyceae red algae dominate many geothermal habitats and provide important tools for investigating the evolution of extremophilic eukaryotes and associated microbial communities. We propose that resource sharing drove genome reduction in Cyanidiophyceae and enabled the neofunctionalization of genes in multi-enzyme pathways. Utilizing arsenic detoxification as a model, we discuss how the sharing of gene functions by other members of the microbial assemblage weakened selection on homologs in the Cyanidiophyceae, allowing long-term gene persistence via the putative gain of novel functions. This hypothesis, referred to as the Integrated Horizontal Gene Transfer (HGT) Model (IHM), attempts more generally to explain how extremophilic eukaryotes may have transitioned from 'hot start' milieus by functional innovations driven by the duplication and divergence of HGT-derived genes.

|
Scooped by
?
Today, 1:26 PM
|
Big Science projects are often troubled by misalignments. We analyze how synthetic cell researchers handle misalignments by performing alignment work. We find that alignment work renders Big Science feasible, but involves trade-offs, for example between short-term successes and long-term goals. We thus recommend attending to the politics of alignment work.

|
Scooped by
?
Today, 1:13 PM
|
Traditionally perceived as an RNA-specific nuclease, Cas13a has been used primarily for RNA detection. We discover the ability of Leptotrichia buccalis Cas13a (LbuCas13a) to directly target DNA without the restrictions of protospacer flanking sequence and PAM sequences, coupled with robust trans-cleavage activity. Contrary to conventional understanding, LbuCas13a does not degrade DNA targets. Our study reveals an enhancement in the single-nucleotide specificity of LbuCas13a against DNA compared to RNA. This heightened specificity is attributed to the lower affinity of CRISPR RNA (crRNA) towards DNA, raising the crRNA–DNA binding energy barrier. We introduce a molecular diagnostic platform called superior universal rapid enhanced specificity test with LbuCas13a (SUREST) for high-resolution genotyping. SUREST is capable of detecting DNA concentrations of CYP2C19 (rs4986893) as minute as 0.3 aM (0.18 cps µl−1). We also apply SUREST to human genotyping scenarios, indicating that SUREST performs well across a broad range of mutations and sequence contexts. SUREST represents an advancement in real-time nucleic acid detection, making it a useful tool for pathogen identification and mutation analysis in clinical diagnostics. A Leptotrichia buccalis Cas13a diagnostic platform can be used to cleave and detect small concentrations of viral and human DNA.

|
Scooped by
?
Today, 1:00 PM
|
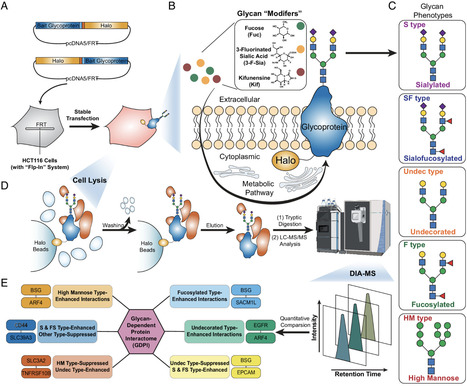
Protein–protein interactions (PPI) are crucial for comprehending the molecular mechanisms and signaling pathways underlying diverse biological processes and disease progression. However, investigating PPIs involving membrane proteins is challenging due to the complexity and heterogeneity of glycosylation. To tackle this challenge, we developed an approach termed glycan-dependent affinity purification coupled with mass spectrometry (GAP–MS), specifically designed to characterize changes in glycoprotein PPIs under varying glycosylation conditions. GAP–MS integrates metabolic control of glycan profiles in cultured cells using small molecules referred to as glycan modifiers with affinity purification followed by mass spectrometry analysis (AP–MS). Here, GAP–MS was applied to characterize and compare the interaction networks under five different glycosylation states for four bait glycoproteins: BSG, CD44, EGFR, and SLC3A2. This analysis identified a network comprising 156 interactions, of which 131 were determined to be glycan dependent. Notably, the GAP–MS analysis of BSG provided distinct information regarding glycosylation-influenced interactions compared to the commonly used glycosylation site mutagenesis approach combined with AP–MS, emphasizing the unique advantages of GAP–MS. Collectively, GAP–MS presents distinct insights over existing methods in elucidating how specific glycosylation forms impact glycoprotein interactions. Additionally, the glycan-dependent interaction networks generated for these four glycoproteins serve as a valuable resource for guiding future functional investigations and therapeutic developments targeting the glycoproteins discussed in this study.

|
Scooped by
?
Today, 1:04 AM
|
Testing microbial interaction hypotheses is a major challenge in microbiome research. Bacteriophages may be used to probe organism-organism interactions within complex microbiomes, yet are rarely used for this purpose. Here, we isolated nine narrow host range phages that replicate in several Variovorax species. As Variovorax CL14 degrades the plant root stunting hormone auxin, we used phages to eliminate V. CL14 from a rhizosphere consortium and reestablished the stunted root phenotype. We used three of the phages to deplete another Variovorax, SCN45, from complex communities to test correlation network-based hypotheses of thiamine interdependencies. Genome-resolved metagenomics revealed that three taxa decreased in relative abundance following Variovorax depletion and could be rescued by thiamine supplementation. Thus, we confirmed thiamine production as the mechanistic basis for interdependence. These experiments lay the foundation for research that employs wildtype and genetically modified phages to test interaction hypotheses and for targeted microbiome manipulation.

|
Scooped by
?
Today, 12:54 AM
|
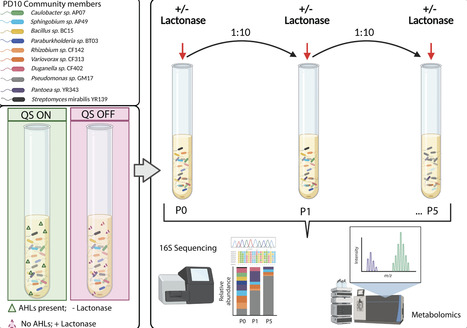
Bacteria are recognized for their diverse metabolic capabilities, yet the impact of microbe-microbe interactions on multispecies community structure and dynamics is poorly understood. Cell-to-cell signaling in the form of quorum sensing (QS) often regulates secondary metabolite production and microbial interactions. Here, we examine how acylhomoserine lactone (AHL)-mediated QS impacts microbial community structure in a 10-member synthetic community of isolates from Populus deltoides. To explore the role of QS in microbial community structure and dynamics, we disrupted AHL signaling by exogenous addition of AiiA-lactonase, an enzyme that cleaves the lactone ring. Microbial community structure resulting from signal inactivation, as measured by 16S rRNA amplicon sequencing and secondary metabolite production, was assessed after successive passaging of the community. Further, we investigated the impact of quorum quenching on specific microbe-microbe interactions using pairwise inhibition assays. Our results indicate that AHL inactivation alters the relative abundance of dominant community members at later passages but does not impact the overall membership in the community. Quorum quenching significantly alters the metabolic profile in lactonase-treated communities. This metabolic alteration impacts microbe-microbe interactions through decreased inhibition of other community members. Together, these results indicate that QS impacts microbial community structure through the regulation of secondary metabolites in dominant members and that the membership of microbial communities can be relatively stable despite changes in metabolic profiles.

|
Scooped by
?
Today, 12:32 AM
|
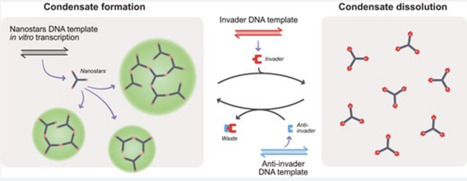
RNA-driven phase separation is emerging as a promising approach for engineering biomolecular condensates with diverse functionalities. Condensates form thanks to weak yet specific RNA–RNA interactions established by design via complementary sequence domains. Here, we demonstrate how RNA condensates formed by star-shaped RNA motifs, or nanostars, can be dynamically controlled when the motifs include additional linear or branch-loop domains that facilitate access of regulatory RNA molecules to the nanostar interaction domains. We show that condensates dissolve in the presence of RNA “invaders” that occlude selected nanostar bonds and reduce the valency of the nanostars, preventing phase separation. We further demonstrate that the introduction of “anti-invader” strands, complementary to the invaders, makes it possible to restore condensate formation. An important aspect of our experiments is that we demonstrate these behaviors in one-pot reactions, where RNA nanostars, invaders, and anti-invaders are simultaneously transcribed in vitro using short DNA templates. Our results lay the groundwork for engineering RNA-based assemblies with tunable, reversible condensation, providing a promising toolkit for synthetic biology applications requiring responsive, self-organizing biomolecular materials.

|
Scooped by
?
Today, 12:17 AM
|
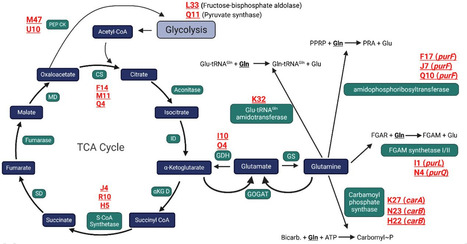
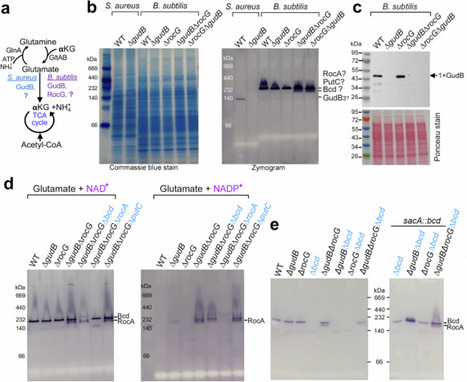
Glutamate dehydrogenase (GDH) resides at the crossroads of nitrogen and carbon metabolism, catalyzing the reversible conversion of L-glutamate to α-ketoglutarate and ammonium. GDH paralogs are ubiquitous across most species, presumably enabling functional specialization and genetic compensation in response to diverse conditions. Staphylococcus aureus harbors a single housekeeping GDH (GudB), whereas Bacillus subtilis encodes both a major and a minor GDH, GudB and RocG, respectively. In an unsuccessful attempt to identify an alternative GDH in S. aureus, we serendipitously discovered previously unrecognized GDH activity in two metabolic enzymes of B. subtilis. The hexameric Val/Leu/Ile dehydrogenase Bcd (formerly YqiT) catabolizes branched-chain amino acids and to a lesser extent glutamate using NAD+ as a cofactor. Removal of gudB and rocG unmasks the dual NAD(P)+-dependent GDH activity of RocA, which otherwise functions as a 3-hydroxy-1-pyrroline-5-carboxylate dehydrogenase. Bcd homologs are prevalent in free-living and obligate bacteria but are absent in most, if not all, staphylococci. Despite low sequence homology, Bcd structurally resembles the GudB/RocG family and can functionally compensate for the loss of GudB in S. aureus. Bcd is essential for the full maturation of biofilms. B. subtilis lacking GDHs exhibits severe impairments in rugose architecture and colony expansion of biofilms. This study underscores the importance of metabolic redundancy and highlights the critical role of substrate promiscuity in GDHs during biofilm development.

|
Scooped by
?
June 21, 5:02 PM
|
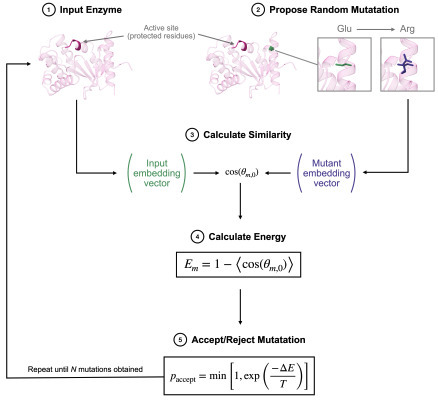
We present a hybrid approach combining a protein language model (pLM) with Monte Carlo (MC) sampling for generating enzyme mutants free of mutations deleterious for structural preservation. Given the amino acid sequence of the original enzyme and a set of residues for which the local environment should be conserved, i.e., the catalytic site, our approach generates mutants that differ vastly in the overall sequence while retaining the geometry of the conserved region, thereby representing promising candidates for further experimental screening. Unlike end-to-end deep learning approaches, whose results are harder to interpret and control, the use of a well-established, classic technique such as MC sampling allows us to easily interpret the generative process as the sampling of an energy landscape determined by the pLM. In turn, such an interpretation enables us to steer this generative process and control its outcome by making use of robust statistical mechanics concepts, e.g., temperature, thereby explicitly guaranteeing certain properties of the generated mutants. Given the increasing relevance of generative algorithms in the design and search for novel, optimised enzymes, we believe that our results constitute an important step for the future development of this class of techniques. To facilitate experimental verification, we finally provide hundreds of sequences for 13 different enzymes involved in catalytic processes ranging from carbon dioxide conversion to DNA replication.

|
Scooped by
?
June 21, 4:55 PM
|
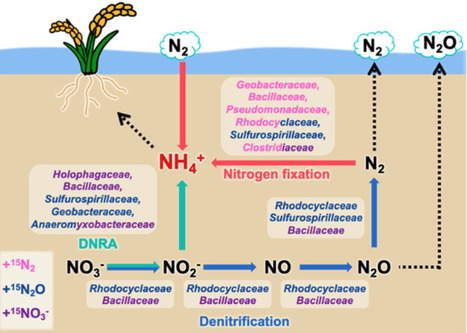
Rice paddy fields are sustainable agricultural systems as soil microorganisms help maintain nitrogen fertility through generating ammonium. In these soils, dissimilatory nitrate reduction to ammonium (DNRA), nitrogen fixation, and denitrification are closely linked. DNRA and denitrification share the same initial steps and nitrogen gas, the end product of denitrification, can serve as a substrate for nitrogen fixation. However, the microorganisms responsible for these three reductive nitrogen transformations, particularly those focused on ammonium generation, have not been comprehensively characterized. In this study, we used stable isotope probing with 15NO3−, 15N2O, and 15N2, combined with 16S rRNA high-throughput sequencing and metatranscriptomics, to identify ammonium-generating microbial consortia in paddy soils. Our results revealed that several bacterial families actively contribute to ammonium generation under different nitrogen substrate conditions. Specifically, Geobacteraceae (N2O and +N2), Bacillaceae (+NO3− and +N2), Rhodocyclaceae (+N2O and +N2), Anaeromyxobacteraceae (+NO3− and +N2O), and Clostridiaceae (+NO3− and +N2) were involved. Many of these bacteria participate in key ecological processes typical of paddy environments, including iron or sulfate reduction and rice straw decomposition. This study revealed the ammonium-generating microbial consortia in paddy soil that contain several key bacterial drivers of multiple reductive nitrogen transformations and suggested their diverse functions in paddy soil metabolism.

|
Scooped by
?
June 21, 4:28 PM
|
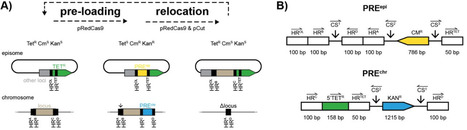
We have developed a simple and highly efficient in vivo method to iteratively relocate functional chromosomal loci onto an episome in Escherichia coli by utilizing synthetic DNA fragments. In this in vivo cut’n’paste procedure, “cutting” is executed by the RNA-guided DNA endonuclease Cas9 and a set of guides, while “pasting” is facilitated by the phage λ Red recombinase, which are all synthesized on easily curable plasmids. To demonstrate the utility of this approach, we commercially obtained synthetic DNA fragments containing locus-specific homology regions, antibiotic marker cassettes, and standardized Cas9 target sequences. By using these locus-specific synthetic DNA fragments, we relocated 7 functional chromosomal loci. Scarless relocation mutants of the trg, aer, tsr, malHM, malQT, macB-nadA, and folA chromosomal loci were obtained as antibiotic-resistant isolates by combining Cas9 counterselection with the restoration of an antibiotic marker cassette. The additional antibiotic marker cassettes and standardized Cas9 target sequences present in the synthetic DNA fragments are inherently eliminated upon completion of the procedure, enabling iterative processing of loci. This procedure should be widely useful, especially in genome (re-)engineering of E. coli and other bacteria because the procedure can be done on wild-type cells.

|
Scooped by
?
June 21, 4:01 PM
|
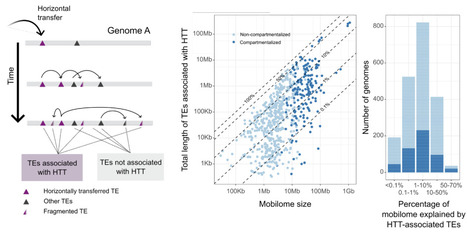
Transposons impact eukaryotic genome size and evolution. Horizontal transfer of transposons (HTT) is important for their long-term persistence, but has only been systematically studied in animals, and thus the abundance, impact, and factors that shape HTTs in lineages outside animals is unknown. Fungi are at least as ancient and diverse as animals and are characterized by extensive genome size variation caused by transposons. Here, we screened 1,348 genomes across fungal biodiversity, genome sizes, and lifestyles to detect extensive HTTs, that generated on average 7% but up to 70% of the transposon content in some taxa. We in total identified at least 5,518 independent HTTs, mostly involving Tc1/Mariner DNA transposons. While the majority of HTTs occur between closely related taxa, irrespective of their lifestyles, HTTs were particularly common in Mucoromycotina, Sordariomycetes, Dothideomycetes, and Leotiomycetes. Importantly, species lacking fungal-specific defense mechanisms against transposons and those with gene-sparse and repeat-rich genomic compartments are involved in significantly higher number of HTTs, unveiling ecological and genomic factors shaping HTTs. Our findings thus illuminate the dynamic landscape of HTTs in fungi, providing the framework to further study the impact of HTTs on genome evolution and the processes that mediate transposon transfers within and between eukaryotic lineages.
|
 Your new post is loading...
Your new post is loading...