 Your new post is loading...

|
Scooped by
?
Today, 11:19 AM
|
Gene mutation is the primary source of genetic variation, yet natural mutation rates are insufficient to meet the demands of biotechnological applications. This review systematically examines advancements in hypermutation technologies designed to overcome this limitation, focusing on targeted, multi-target, and genome-wide approaches, and their transformative applications across diverse fields. By comparing these tools across multiple dimensions, including mutation scope, rate, and type, we provide insights to guide the selection of optimal mutagenesis tools for accessing distinct genetic landscapes and addressing practical challenges. Furthermore, we summarize the challenges that persist in this active area and highlight future directions such as developing high-throughput screening methods and AI-driven predictive models for mutational outcomes. By bridging the gap between natural mutation constraints and biotechnological needs, hypermutation technologies promise to unlock unprecedented innovations in synthetic biology, evolutionary research, and industrial applications and pave the way for exploring previously inaccessible genetic landscapes.

|
Scooped by
?
Today, 11:06 AM
|
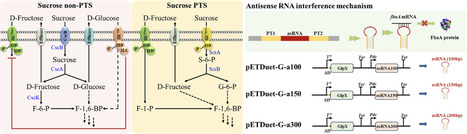
d-Allulose, a rare sugar characterized by its high sweetness and low-calorie profile, is gaining attention in the sweetener market. This study introduces an innovative method for converting sucrose into d-allulose through microbial fermentation. An irreversible synthesis pathway was constructed by expressing the scrA, scrB, alsE, and a6PP genes in Escherichia coli JM109 (DE3), enhancing substrate utilization via dual PTS-dependent transport of sucrose and d-fructose. A fructose-1,6-bisphosphatase mutant (GlpX [K29A]) was used to facilitate the influx of fructose-1-phosphate into the synthesis pathway. The Embden–Meyerhof–Parnas (EMP) and pentose phosphate (PP) pathways were weakened by deleting the pfkA and rpiA genes. To further regulate carbon fluxes, a structurally stable antisense RNA (asRNA) was employed to inhibit FbaA expression. The fermentation medium was optimized using response surface methodology. Finally, the d-allulose titer reached 12.8 g/L, with a yield of 0.23 g/g on sucrose, achieved through fed-batch fermentation in a 5 L fermenter.

|
Scooped by
?
Today, 10:23 AM
|
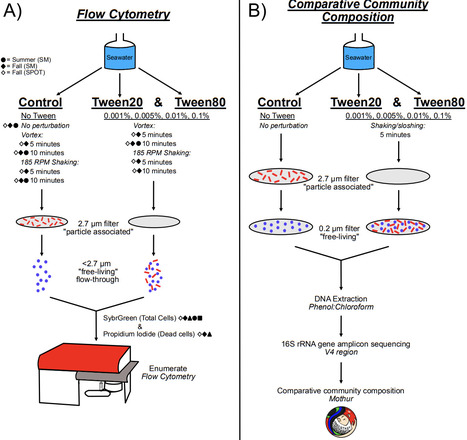
Marine particles, typically composed of organic detritus and cellular debris, harbor microbial communities that are distinct from the planktonic, or free-living, communities in the pelagic ocean. However, without being first separated from the particle and microbial consortia, these microbes are inaccessible to further investigation via single-cell microbiology methods like flow cytometry, cell-sorting, and dilution-based isolation. To confront this obstacle, we compared the dissociative effects of two commonly used detergents, Tween20 and Tween80, on particle-associated marine microbial communities. The ability of Tween treatments to liberate cells from particles, and to maintain cell integrity, was quantified by flow cytometry from multiple communities across seasons and locations. Both Tween20 and Tween80, at 185 RPM shaking, gently dissociated microbes from their particles, causing very little cell mortality. Additionally, Tween80 liberated a greater number of particle-associated cells into the free-living fraction. We also analyzed the effects of Tween treatments on the microbial community composition for one of these collections via 16S rRNA gene amplicon sequencing of the particle-associated and free-living fractions relative to unamended controls. Tween20 and Tween80 were both effective for microbial dissolution from particles, however Tween80 treatments displayed greater uniformity in the dissociated communities and significantly enriched for the most abundant particle-associated members. Together these data indicate that Tween80 was most effective at gently dissociating particle-associated cells.

|
Scooped by
?
Today, 9:51 AM
|
Plasmids constitute a key tool in synthetic biology, providing a versatile framework for various research and industrial ventures. An essential determinant of plasmid functionality is its copy number, impacting both protein production rates and host cell metabolic burden. However, currently there is no model that can computationally predict the plasmid copy number from the sequence of its origin of replication (ORI). We present a novel software solution tailored to simplify plasmid copy number design, poised to redefine plasmid engineering workflows. At the heart of our tool lies a comprehensive machine learning model, informed by numerous features extracted from the ORI. This computational model emphasizes the importance of promoter strength and the RNA folding dynamics of regulatory elements within the ORI. Additionally, we detail a robust protocol for the efficient manipulation of plasmid ORIs used to validate our model’s predictive capabilities. This innovation represents a paradigm shift in plasmid-centric methodologies, offering unprecedented avenues for advancement in synthetic biology research and industrial applications. The software is available at: https://pcn-gradient.vercel.app/

|
Scooped by
?
Today, 9:24 AM
|
Weber et al. (2025; doi: 10.1111/nph.70161) importantly advances our understanding of the relationship between AM fungal biodiversity and plant productivity. Employing an impressive experimental design, including 37 AM fungal communities of variable richness and double isotopic labelling, the authors demonstrated a direct relationship between AM fungal species richness and phosphorus supply to plants via the mycorrhizal pathway. Furthermore, with carbon flow to fungal hyphae remaining constant along the fungal diversity gradient, higher AMF species richness made the exchange rate of the two key resources more favorable to the plants.

|
Scooped by
?
Today, 1:13 AM
|
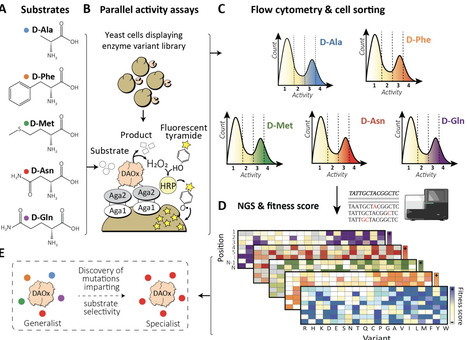
Substrate specificity is a defining feature of enzyme function, but its molecular underpinnings remain difficult to decode and engineer. Here, we leveraged enzyme proximity sequencing (EP-Seq) to systematically map how single-point and combinatorial mutations reshape the substrate preferences of D-amino acid oxidase (DAOx) from Rhodotorula gracilis, a model promiscuous enzyme. We generated ~40,000 sequence-phenotype pairs, enabling us to profile the activities of ~6,500 unique DAOx variants against five D-amino acid substrates with distinct physicochemical properties. Our analysis revealed that substrate-specific mutations are distributed throughout the enzyme structure. Mutations near the active site drive strong specificity shifts but also incur catalytic penalties, while distal mutations subtly rewire intramolecular contacts in order to modulate specificity with minimal loss of activity. We identified and validated positional hotspots that act allosterically to influence specificity, and characterized key variants that acquired exclusive substrate specificity or exhibited up to 230-fold changes in substrate preference. Combining mutations with complementary effects further sharpened substrate discrimination, enabling rational design of highly selective biocatalysts. This work provides a powerful framework for decoding enzyme specificity and provides unique foundational datasets to advance AI-guided enzyme engineering.

|
Scooped by
?
Today, 12:11 AM
|
Adaptation to an environment is enabled by the accumulation of beneficial mutations. How do adaptive trajectories and pleiotropic effects of adaptation change in response to “subtle” changes in the environment? Since there exists no molecular framework to quantify “subtle” environmental change, designing experiments to answer this question has been challenging. In this work, we address this question by studying the effects of evolution in environments which differ solely in the way sugars are presented to a bacterial population. Specifically, we focus on glucose and galactose, which can be supplied to an E. coli population as a mixture of glucose and galactose, lactose, or melibiose. We evolve six replicate populations of E coli for 300 generations in these three chemically correlated or “synonymous” environments, and show that the adaptive responses of these populations are not similar. When tested for pleiotropic effects of fitness in a range of non-synonymous environments, our results show that despite uncorrelated adaptive changes, the nature of pleiotropic effects is largely predictable based on the fitness of the ancestor in the non-home environments. Overall, our results highlight how subtle changes in the environment can alter adaptation, but despite sequence-level variations, pleiotropy is qualitatively predictable.

|
Scooped by
?
July 15, 11:54 PM
|
The widespread use of glyphosate has led to a rapid increase in glyphosate resistant weeds at a considerable cost to agriculture. Furthermore, agricultural run-off means non-target, natural ecosystems are also affected. Resistance may arise both through mutations to target enzyme EPSPS in the shikimate pathway generating insensitivity to glyphosate and through non-target site mutations regulating the dose of glyphosate reaching the shikimate pathway. Either mechanism may trade-off against normal cell functioning. Here, using experimental evolution of replicated Chlamydomonas reinhardtii populations facing glyphosate, we reveal the metabolomic fingerprint of both glyphosate action and emerging resistance by applying untargeted metabolomic screens throughout the course of resistance evolution. Furthermore, this allows us to evaluate potential underlying molecular mechanisms and fitness costs. We find evidence of build-up of shikimate pathway metabolites, a characteristic signal of glyphosate action, that subsides but does not disappear as resistance evolves, suggesting a fitness trade-off. Concurrent with this is evidence of cell wide disruption of the amino acid pool, that stabilizes as resistance evolves. We also found evidence of effects of glyphosate on membrane lipids and increased levels of reactive oxygen species persisting after resistance had evolved. This suggests a considerable effect of the recently described secondary effect of glyphosate: oxidative damage. These data highlight several metabolic processes disrupted by and persisting with the evolution of resistance to glyphosate and may provide a template for enhancing predictions of population and ecosystem effects.

|
Scooped by
?
July 15, 11:45 PM
|
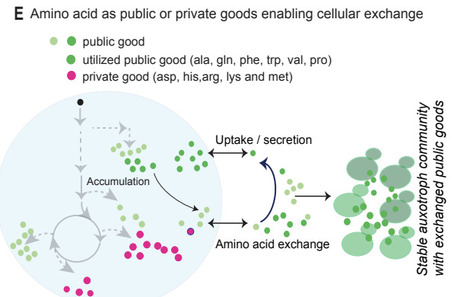
The biosynthetic capacity of the cell governs the production and exchange of amino acids. Amino acids have diverse metabolic origins and intracellular requirements. Therefore, to understand the cellular amino acid economy we require a quantitative understanding of intracellular and extracellular their amounts, and how distinct amino acids change temporally during growth. Using the unicellular eukaryote, yeast, we establish a quantitative blueprint of the intracellular and extracellular amino acid economy, including the fluxes of production, secretion and consumption across different growth phases. Certain amino acids dominate the intracellular pool, and proportions of distinct amino acids continuously change during growth. The extracellular pool is notably distinct from the intracellular pool in terms of composition and dynamics. Only some public amino acids are subsequently consumed, while others are secreted in surplus of cellular demands. Five amino acids are intracellular and privatized, and continuously utilized to support diverse metabolism. Through this, we establish pairs of highly effective synthetic, exchange-based communities of public good auxotrophs. Our results suggest organizing frameworks addressing the dynamic intracellular and inter-cellular amino acid economy and its trade, and informs the rational engineering of syntrophic cell communities.

|
Scooped by
?
July 15, 10:19 PM
|
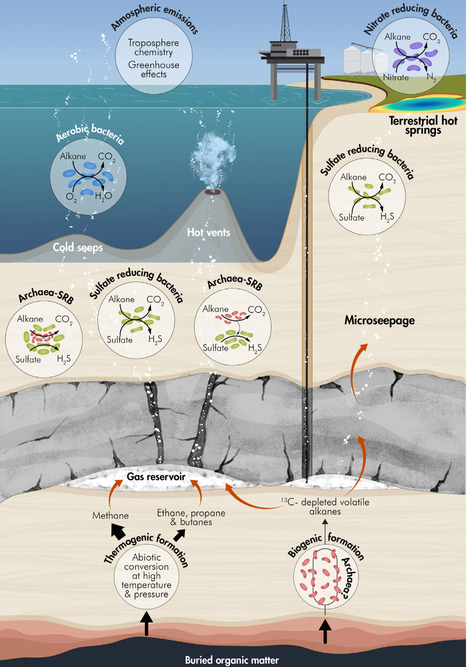
The short-chain volatile alkanes ethane, propane, and butane are major components of natural gas. Released from deep-seated subsurface reservoirs through natural seepage or gas extraction, they percolate through anoxic and oxic environments before reaching the atmosphere, where they contribute to tropospheric chemistry and act as greenhouse gases. While their aerobic biological oxidation is well established, their fate in anoxic environments has only recently come into focus. Here we review their oxidation in anoxic settings – from subsurface reservoirs and deep-sea seep sediments to terrestrial hot springs and wastewater treatment plants. We discuss the phylogenetic diversity, biochemical mechanisms, and physiology of microorganisms mediating anaerobic oxidation of volatile alkanes, including nitrate- and sulfate-reducing bacteria (SRB) and the recently discovered alkane-oxidizing archaea. We also highlight advances in diagnostic tools, such as stable isotope analyses and single-cell chemical imaging. Finally, we outline major unresolved research questions, including the unique biochemistry of anaerobes and the extent to which they act as natural biofilters by reducing atmospheric emissions of volatile alkanes.

|
Scooped by
?
July 15, 4:30 PM
|
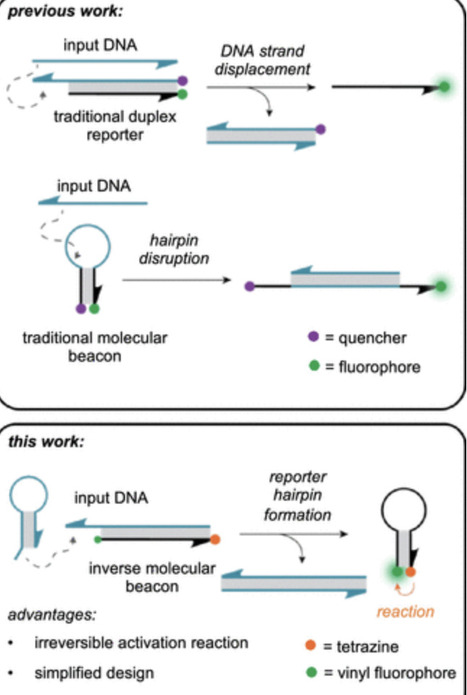
Nucleic acid–based hybridization probes that produce a fluorescent signal in the presence of DNA or RNA target molecules are essential components of nucleic acid computation and detection strategies. Commonly, the fluorescence activation of reporter gates is triggered by separation of a fluorophore–quencher pair upon target hybridization or strand displacement. In order to expand the utility of DNA computing by providing a chemical reaction as the ultimate output, reporter systems have been designed that carry reactive groups, which undergo a proximity-induced reaction upon oligonucleotide hybridization. The downside of published reporter gate designs is that they are composed of two separate, chemically modified oligonucleotides, which need to be taken into consideration when designing upstream circuits. Here, we report a novel hairpin-forming nucleic acid reporter probe that utilizes template-induced proximal reactivity to activate a small molecule in the presence of an unmodified nucleic acid input molecule. This DNA hairpin reporter gate consists of a duplex between a blocking strand and a hairpin-forming reporter strand. In the presence of input, the blocking strand is displaced, triggering hairpin formation allowing the proximity-driven templated activation of a vinyl ether-caged fluorophore by a tetrazine via an inverse electron demand Diels–Alder reaction. This new approach demonstrates robust small-molecule activation in vitro and in cells through logic operations in the presence of input DNA molecules.

|
Scooped by
?
July 15, 12:52 PM
|
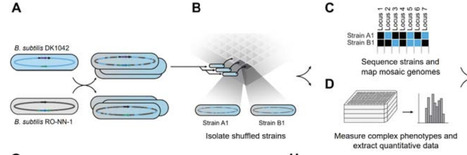
Genetic mapping is a powerful tool for eukaryotic genetics that has only been applied to bacteria in limited circumstances. Quantitative trait locus (QTL) mapping generally relies on sexual recombination to break linkages between genes, yet bacteria rarely undergo sufficient homologous recombination to generate suitable mapping populations. In this work, we used iterative biparental genome shuffling by protoplast fusion in Bacillus subtilis to generate a population of bacteria with substantial random recombination throughout their genomes. Individual shuffled progeny were arrayed in well plates, resequenced, and characterized for a range of complex phenotypes including spore germination and swarming motility. Genetic mapping of the resulting phenotypes identified high-confidence QTLs of moderate size (~10 kb), and these associations were validated through targeted genetic swaps. This B. subtilis QTL population can easily be used to map additional phenotypes and the general approach for QTL mapping is applicable in a wide range of bacteria.

|
Scooped by
?
July 15, 12:34 PM
|
Automated protein function prediction/annotation (AFP) is vital for understanding biological processes and advancing biomedical research. Existing text-based AFP methods including the state-of-the-art method, GORetriever, rely on expert-curated relevant literature, which is costly and time-consuming, and cover only a small portion of the proteins in UniProt. To overcome this limitation, we propose GOAnnotator, a novel framework for automated protein function annotation. It consists of two key modules: PubRetriever, a hybrid system for retrieving and re-ranking relevant literature, and GORetriever+, an enhanced module for identifying Gene Ontology (GO) terms from the retrieved texts. Extensive experiments over three benchmark datasets demonstrate that GOAnnotator delivers high-quality functional annotations, surpassing GORetriever in realistic situations by uncovering unique literature and predicting additional functions. These results highlight its great potential to streamline and enhance annotation of protein functions without relying on manual curation.
|

|
Scooped by
?
Today, 11:14 AM
|
Indigoidine, a natural bio-pigment with a planar conjugated structure comprising two pyrrolidinone rings linked by a central double bond, exhibits unique chromogenic properties. Growing environmental concerns over synthetic and plant-derived pigments have driven interest in microbial pigments as sustainable alternatives. Despite the potential of nonribosomal peptide synthetases (NRPSs) to optimize indigoidine biosynthesis, mechanistic insights into their catalytic functions remain limited. This review systematically outlines the discovery and evolution of indigoidine, focusing on the structural features of NRPSs and associated biosynthetic pathways. It evaluates current strategies for enhancing production, including host engineering, metabolic flux optimization, genome-wide screening, transcriptional regulation, and scale-up processes. Additionally, the applications of indigoidine in textile, agricultural, and biomedical industries are critically examined. By proposing a novel research framework for microbial pigments synthesis optimization, this review contributes theoretical and technical advancements toward sustainable biopigment manufacturing.

|
Scooped by
?
Today, 10:32 AM
|
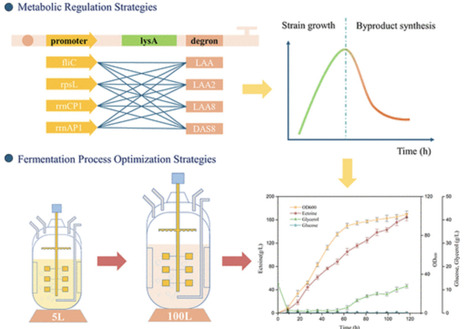
Ectoine is a pivotal natural osmoprotectant that functions as a compatible solute through osmoregulation, enabling microorganisms to thrive in extreme environments such as high salinity. To meet market demands, this study focuses on optimizing its production process. We initially engineered the ectABC gene cluster from Halomonas venusta via 5′-UTR modification, establishing a functional ectoine biosynthesis pathway in E. coli. Subsequent introduction of a rate-limiting enzyme EctB mutant (E407D) and aspartokinase mutant increased titer by 140%. To address lysine byproduct accumulation, an innovative molecular switch was employed to regulate lysA gene expression, achieving dynamic balance between cell growth and product synthesis. Further optimization through cofactor engineering yielded the final strain ECT31, which produced 164.6 g/L ectoine in a 100 L bioreactor within 117 h, the highest reported titer for E. coli-based ectoine production to date. The metabolic engineering strategy presented herein establishes a new pdigm for efficient biosynthesis of amino acid derivatives.

|
Scooped by
?
Today, 10:02 AM
|
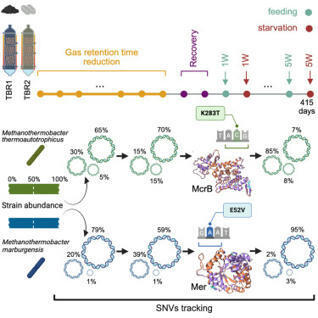
Genetic heterogeneity exists within all microbial populations, with sympatric cells of the same species often exhibiting single-nucleotide variations that influence phenotypic traits, including metabolic efficiency. However, the evolutionary dynamics of these strain-level differences in response to environmental stress remain poorly understood. Here, we present a first-of-its-kind study tracking the adaptive evolution of an anaerobic, carbon-fixing microbiota under a controlled engineered ecosystem focused on carbon dioxide bioconversion into methane. Leveraging strain-resolved metagenomics with an ad hoc variant calling and phasing approach, we mapped mutation trajectories and observed that the two dominant Methanothermobacter species maintained distinct sweeping haplotypes over time, most likely due to niche-specific metabolic roles. By combining population genetic statistics and peptide reconstruction, mer and mcrB genes emerged as potential drivers of archaeal strain-level competition. These findings pave the way for targeted engineering of microbial communities to enhance bioconversion efficiency, with significant implications for sustainable energy and carbon management in anaerobic systems.

|
Scooped by
?
Today, 9:48 AM
|
Bifidobacteria are beneficial saccharolytic microbes that are widely used as probiotics or in synbiotic formulations, yet individual responses to supplementation can vary with strain type, microbiota composition, diet and lifestyle, underscoring the need for strain-level insights into glycan metabolism. Here we reconstructed 68 pathways for the utilization of mono-, di-, oligo- and polysaccharides by analysing the distribution of 589 curated metabolic gene functions (catabolic enzymes, transporters and transcriptional regulators) across 3,083 non-redundant Bifidobacterium genomes of human origin. Thirty-eight predicted phenotypes were validated in vitro for 30 geographically diverse strains, supporting genomics-based predictions. Our analysis uncovered extensive inter- and intraspecies functional heterogeneity, including a distinct clade within Bifidobacterium longum that metabolizes α-glucans and Bangladeshi isolates carrying unique gene clusters for xyloglucan and human milk oligosaccharide utilization. This large-scale genomic compendium advances our understanding of bifidobacterial carbohydrate metabolism and can inform the rational design of probiotic and synbiotic formulations tailored to strain-specific nutrient preferences. A comprehensive genomic analysis reveals species- and strain-level heterogeneity in carbohydrate utilization potential across bifidobacteria of human origin.

|
Scooped by
?
Today, 9:20 AM
|
Open compartments, termed biomolecular condensates, are involved in cellular reactions. This Comment highlights their ability to enhance robustness and control of reactions in space and time, going beyond mere acceleration or inhibition. They enable reaction engineering at the mesoscale, not just by altering concentrations but also by modifying the local environment.

|
Scooped by
?
Today, 12:30 AM
|
While extracellular vesicles (EVs) are established mediators of intra-species signaling, their role as active participants in cross-kingdom communication remains incompletely understood. Here, we reveal that human colon cells and both Gram-positive and Gram-negative gut bacteria engage in species-specific, EV-mediated molecular dialogue, driven in part by RNA cargo. We show that bacterial EVs (BEVs) induce distinct transcriptomic responses in human cells, and that BEV-RNA independently causes similar effects. Conversely, we demonstrate that human EVs and highly abundant miR-192-5p are differentially internalized by bacteria, affecting their physiology. Our findings support a conceptual model in which EVs function as directional messengers that shape host-microbiome interactions. This study introduces a framework for understanding EVs as cross-kingdom regulators and underscores the importance of tailored, context-specific analyses for understanding the scope of EV-mediated interactions in microbiome-host homeostasis and disease.

|
Scooped by
?
Today, 12:07 AM
|
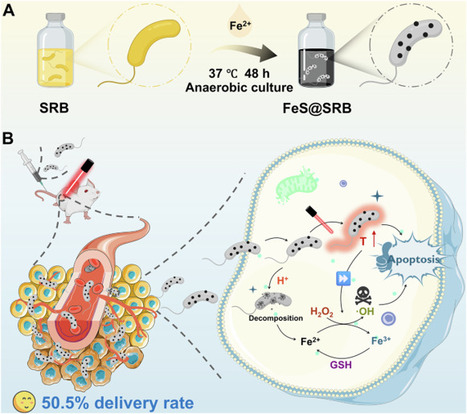
Anaerobic bacteria-mediated tumor therapy faces multiple challenges, including potential toxic side effects, complex manufacturing processes, and impaired hypoxic targeting. Here, based on the excellent biocompatibility and distinctive metabolic ability of natural anaerobic sulfate-reducing bacteria (SRB) to dissimilate sulfate into sulfide, we construct in situ-biosynthesized ferrous sulfide nanoparticle-SRB (FeS@SRB) biohybrid to enhance tumor-targeted therapy. Interestingly, SRB acts as both a biosynthesis factory and active tumor-targeted delivery vehicles. Our systematic studies reveal that FeS@SRB has excellent biosafety and tumor targeting capabilities, achieving over 50% tumor delivery efficiency in female mice post-intravenous injection, which is 17 times higher than that of conventional chemically-synthesized FeS@BSA nanoparticles. Upon near-infrared laser irradiation, FeS@SRB hybrids exhibit synergistic photothermal-chemodynamic effect, amplifying oxidative stress to trigger tumor cells ferroptosis and apoptosis, thereby effectively suppressing both subcutaneous and orthotopic tumor growth. This SRB-based therapeutic strategy expands research into tumor-targeting platforms and the biosynthesis of metal sulfide nanoparticles for enhanced tumor therapy. Anaerobic bacteria-mediated tumor therapy has potential but is hindered by complex manufacturing processes, impaired targeting efficiency and side effects. Here, authors develop non-pathogenic anaerobic sulfate-reducing bacteria for in situ biosynthesis of ferrous sulfide nanoparticles for tumor-targeted therapy.

|
Scooped by
?
July 15, 11:47 PM
|
The Bacillus genus contains many members with food significance, including the food-grade Bacillus subtilis clade often used in fermentations and the pathogenic Bacillus cereus clade. Chemically defined media for Bacillus species are crucial tools to allow detailed investigations of the influence of specific nutrients on growth and also improve reproducibility and consistency of experiments. Previous studies have focused on the development of defined media for single species, while the aim of this study was to develop a chemically defined medium that supports the growth of multiple food relevant Bacillus species. The new medium, Pafoba, was tested using two pathogenic strains of the Bacillus cereus clade and eleven strains of the Bacillus subtilis clade representing seven dif-ferent clade members. All thirteen Bacillus strains were able to grow on Pafoba, of which ten dis-played a similar or higher maximum OD600 on Pafoba medium compared to rich medium (Brain Heart Infusion broth). Detailed analysis revealed a biotin requirement for Bacillus subtilis strain PRO64, and the necessity of including essential amino acids for Bacillus weihenstephanensis and Bacillus cereus strains. In conclusion, the chemically defined Pafoba medium provides a controlled and reproducible growth environment for fundamental studies and is suitable for detection and enumeration of a broad range of Bacillus spp. related to food processing and safety.

|
Scooped by
?
July 15, 10:43 PM
|
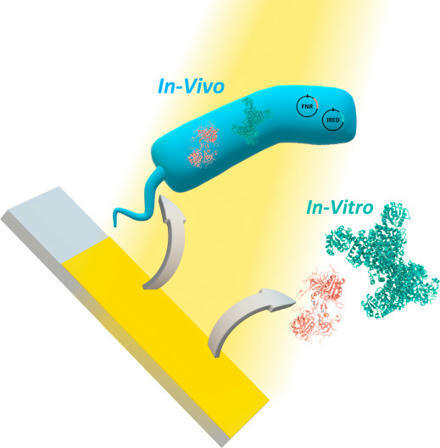
Electrosynthesis is an emerging research direction for greener and more efficient chemical synthesis. Although heterogeneous catalysis efficiency can be improved by tuning electrode surface properties, electrocatalysts frequently fall short of producing chiral molecules with high purity and minimized side reactions. Enzymes are superior catalysts with lower activation barriers. These catalysts, developed through evolution, enable high selectivity and specificity, which are essential for many industrial and pharmaceutical processes. Thus, electronically coupling enzymes or bacteria with electrodes can drive efficient chemical synthesis while ensuring the required selectivity. Here, we used an enzymatic cascade or engineered bacteria for the conversion of 2-methylpyrroline to (R)-2-methylpyrrolidine by isolating or overexpressing imine reductase (IRED), respectively. We further show that coupling the bioelectrocatalytic process with a CdS/NiO-based photoanode enables light-driven, bias-free photo(bio)electrochemical cell activation. The developed platform is versatile and adaptable for any process requiring NADPH-dependent enzymes, in vivo or in vitro.

|
Scooped by
?
July 15, 4:32 PM
|
Cyanobacteria are prolific producers of biologically active compounds that are important in influencing ecology, behavior of interacting organisms, and as leads in drug discovery efforts. Here we discuss the challenges faced by all natural product researchers, especially those that focus on cyanobacteria, and then describe progress that has been made in these areas. We also propose some solutions, paths forward, and thoughts for consideration on these challenges.

|
Scooped by
?
July 15, 4:27 PM
|
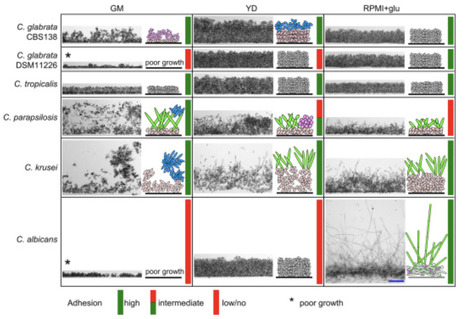
The ability of yeast cells to adhere to solid surfaces or even penetrate semi-solid surfaces and form multicellular biofilms are critical factors in infection. This study examines the relationship between cell adhesion capability and the ability to create spatially organized biofilms in selected Saccharomyces cerevisiae strains, including clinical isolates, and five Candida species (C. albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis). We assessed cell adhesion to polystyrene surface in four media varying in source of carbon and other nutrients. Using microscopy of vertical cell arrangement profiles within yeast populations grown at the solid-liquid interface, we evaluated their internal organization to determine whether the populations exhibit typical biofilm characteristics, such as the spatial organization of distinct cell types. Results indicate that well adherent S. cerevisiae strains form spatial biofilms with typical internal organization, highlighting strain-specific responses to media composition and supporting the use of natural S. cerevisiae strains for biofilm research. Among Candida species, biofilm formation did not consistently align with adhesion efficiency to plastic; while C. albicans and C. krusei formed spatially structured biofilms on media where they adhered well, C. tropicalis and C. glabrata exhibited efficient adhesion without biofilm structuring. Interestingly, C. parapsilosis formed a structured biofilm despite minimal adhesion. These findings emphasize the role of media composition, particularly components of yeast extract and defined medium for mammalian cell growth RPMI, which differentially impacted adhesion and biofilm formation in S. cerevisiae and C. albicans.

|
Scooped by
?
July 15, 12:40 PM
|
An Australian startup from Perth is using seaweed and salt-loving microorganisms to make bio-nylon. Uluu co-founders Julia Reisser and Michael Kingsbury have come up with a properly compostable plastic by feeding red algae to extremophile microbes found in a salt lake in Italy. The microbes, known as halophiles, ferment the sugars from Gracilaria seaweed into polyhydroxyalkanoates (PHAs), a relatively new type of bioplastic that is becoming popular for its ability to fully biodegrade in natural environments. The Uluu team extracts the PHAs and transforms them into pellets, which can be used to make marine-friendly plastic for packaging or textiles. According to the International Union for the Conservation of Nature, 12.7 million metric tonnes of plastic enter the ocean annually. Precedence Research predicts that the biopolymer market will reach $52.3 billion by 2034. Uluu is not the only company to see the potential of seaweed for producing bioplastics. In 2019, researchers from Tel Aviv University fed sea lettuce (Ulva lactuca) to Haloferax mediterranei microbes to create PHAs. Then, in 2020, FlexSea, a biomaterials company from Wolverhampton in the UK, created biopolymers from microbes fed with red seaweed. Uluu is now in the middle of a series A raise and plans to use the funds for a demo plant in Indonesia in 2026.
|
 Your new post is loading...
Your new post is loading...
















2st, we used the recently described mASR (37).