Your new post is loading...

|
Scooped by
?
Today, 1:49 AM
|
Human lactoferrin (HLF), a pivotal iron-binding glycoprotein belonging to the transferrin family, is the primary immune protein in breast milk. Currently, recombinant HLF expression in microorganisms has attracted widespread attention. Through genome integration and copy number optimization, this study established a Komagataella phaffii cell factory that achieved an HLF titer of 137.6 mg/L. However, HLF accumulated extensively within the cells. Subsequently, several endogenous signal peptides were screened and hybridized with mating factor α, resulting in a 1.56-fold increase in extracellular HLF titers compared to the control. Additionally, protein kinase II overexpression facilitated vesicle trafficking, further increasing extracellular HLF to a titer of 304.6 mg/L in shake flasks and 1.7 g/L in a 3-L bioreactor. Notably, the intracellular/extracellular HLF concentration ratio decreased from 1.69:1 to 1.01:1. Further enhancements were made through ion optimization and an exponential methanol-feeding strategy, resulting in an extracellular HLF titer of 3.1 g/L. To the best of our knowledge, this is the first study to report considerably improved extracellular HLF production through optimization of the transport and secretion pathways and provide insights into the expression of other secretary human proteins in K. phaffii.

|
Scooped by
?
Today, 1:41 AM
|
Recent research has indicated the presence of highly protein occupied, transcriptionally silent regions of bacterial genomes which show functional parallels to eukaryotic heterochromatin. We utilized an integrative approach to track chromatin structure and transcription in Escherichia coli K-12 across a wide range of nutrient conditions. In the process, we identified multiple loci which act similarly to facultative heterochromatin in eukaryotes, normally silenced but permitting expression of genes under specific conditions. We also found a strong enrichment of small regulatory RNAs (sRNAs) among the set of differentially expressed transcripts during nutrient stress. Using a newly developed bioinformatic pipeline, the transcription factors (TFs) regulating sRNA expression were bioinformatically predicted, with experimental follow-up revealing novel relationships for 45 sRNA–TF candidates. Direct regulation of sRNA expression was confirmed by mutational analysis for five sRNAs of metabolic interest: IsrB (also known as AzuCR), CsrB and CsrC, GcvB, and GadY. Our integrative analysis thus reveals additional layers of complexity in the nutrient stress response in E. coli and provides a framework for revealing similar poorly understood regulatory logic in other organisms.

|
Scooped by
?
Today, 1:14 AM
|
CRISPR–Cas12a enzymes are versatile RNA-guided genome-editing tools with applications encompassing viral diagnosis, agriculture, and human therapeutics. However, their dependence on a 5′-TTTV-3′ protospacer adjacent motif (PAM) next to DNA target sequences restricts Cas12a’s gene targeting capability to only ∼1% of a typical genome. To mitigate this constraint, we used a bacterial-based directed evolution assay combined with rational engineering to identify variants of Lachnospiraceae bacterium Cas12a with expanded PAM recognition. The resulting Cas12a variants use a range of noncanonical PAMs while retaining recognition of the canonical 5′-TTTV-3′ PAM. In particular, biochemical and cell-based assays show that the variant Flex-Cas12a utilizes 5′-NYHV-3′ PAMs that expand DNA recognition sites to ∼25% of the human genome. With enhanced targeting versatility, Flex-Cas12a unlocks access to previously inaccessible genomic loci, providing new opportunities for both therapeutic and agricultural genome engineering.

|
Scooped by
?
Today, 12:50 AM
|
In this Journal Club, Jian Shu recalls a 2006 publication by Takahashi and Yamanaka as well as a 2021 paper introducing AlphaFold to discuss the fascinating potential of cellular reprogramming in the age of artificial intelligence.

|
Scooped by
?
Today, 12:46 AM
|
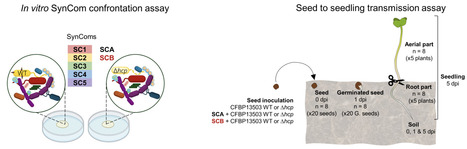
Seeds harbor diverse microbial communities important for plant growth and health. During germination, seed exudation triggers intense microbial competition, shaping the communities transmitted to seedlings. This study explores the role of the bacterial type VI secretion system (T6SS)-mediated interference competition in seed microbiota transmission to seedlings. The analysis of T6SS distribution within 180 genome sequences of seed-borne bacterial strains enabled the construction of synthetic communities (SynCom) with different levels of phylogenetic diversity and T6SS richness. These SynComs were inoculated with Stenotrophomonas rhizophila CFBP13503, a bacterial strain possessing an active T6SS in vitro and in planta. The impact of the T6SS on SynCom composition was assessed in vitro by comparing the CFBP13503 wild-type strain or its isogenic T6SS-deficient mutant co-inoculation. Additionally, the effects of the T6SS on bacterial community dynamics during seed-to-seedling transmission were examined following seed inoculation. The T6SS of S. rhizophila CFBP13503 targets a broad range of bacteria belonging to five different orders. The susceptibility of competing bacteria was partly explained by their phylogenetic proximity and metabolic overlap with CFBP13503. Furthermore, the T6SS modulates the relative abundance of specific bacterial taxa during seed-to-seedling transmission depending on the initial seed inoculum and plant developmental stage. Depending on the sensitivity of the co-inoculated competitors, the T6SS can provide a competitive advantage to CFBP13503, resulting in an increase in population size.

|
Scooped by
?
Today, 12:01 AM
|
Synthetic biology in plants promises to transform basic and applied research by rewiring entire developmental modules, signaling cascades or metabolic pathways. Yet, this requires expression of many genes simultaneously, very difficult with classic transgenic approaches, especially for the generation of stable traits. CRISPR activation systems work in plants and could greatly facilitate multiplexed gene activation. Current CRISPR activation systems are efficient for transient or ubiquitous expression. Yet, to fulfill their potential, CRISPR activation needs to perform robustly in specific organs and tissue types. Here, we present a CRISPRa system that efficiently drives expression in a cell-type-specific manner in stable lines, which requires assessing expression on a cellular basis using fluorescent reporter lines. Our CRISPR systems consistently re-wire gene expression at the cellular level, inducing genes with cell-type specific expression to efficiently express in a new cell layer, such as root endodermis or epidermis. We demonstrate the power of our system to drive functionally relevant, multiplexed gene activation by achieving endodermis-specific production of wild-type levels of flavonoids, detectable by in-situ fluorescence, in a root-flavonoid deficient myb12 mutant. CRISPR activation enables multiplexed, cell-type-specific and stable gene expression in plants. Houbaert et al. successfully reprogrammed gene expression in the root endodermis and restored flavonoid production in mutants, displaying the power of this synthetic biology tool.

|
Scooped by
?
July 16, 11:00 PM
|
Here, we present a major advance of the OrthoFinder method. This extends OrthoFinder's high accuracy comparative genomic framework to provide substantially enhanced scalability and accuracy. Specifically, we show that enhanced phylogenetic delineation of orthogroups provides a 7% relative increase in orthogroup inference accuracy. We further demonstrate that a new gene assignment method substantially reduces overall runtime RAM usage without compromising accuracy. The latest version of OrthoFinder is available at https://github.com/OrthoFinder/OrthoFinder.

|
Scooped by
?
July 16, 10:54 PM
|
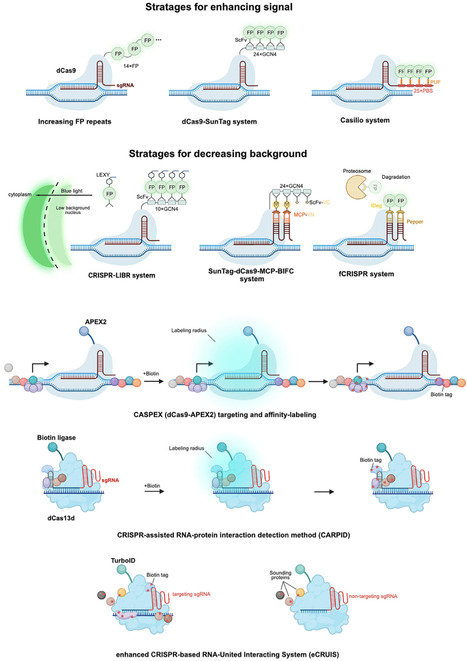
CRISPR technologies have rapidly evolved beyond genome editing, transforming fields such as molecular diagnostics, biosensing, transcriptional regulation, molecular imaging, protein interaction mapping, and single-cell analysis. Emerging CRISPR-based diagnostics harness the collateral cleavage activity of CRISPR-associated (Cas) enzymes for rapid nucleic acid detection. Advanced biosensors extend CRISPR’s capabilities to detect ions, metabolites, and proteins by integrating synthetic biology components. Catalytically inactive Cas proteins enable precise gene regulation and live-cell imaging of nucleic acids, whereas CRISPR-guided proximity labeling has revolutionized the mapping of biomolecular interactions. Recent single-cell CRISPR screens provide unprecedented resolution of cellular heterogeneity. Future research will focus on overcoming current limitations. The integration of CRISPR technologies with artificial intelligence (AI), spatial omics, and microfluidics is expected to further amplify their impact.

|
Scooped by
?
July 16, 8:54 PM
|
Carbon capture is an important mitigation strategy for the reduction of carbon in the atmosphere. Microalgae can simultaneously capture CO2 and valorize biomass, turning waste streams into sustainable bioenergy solutions. However, microalgae struggle when CO2 is either too scarce or too abundant. The scalability of microalgal carbon capture requires the development of strains that can sustain productivity and carbon conversion under fluctuating CO2 levels. In this review, we reframe CO2 assimilation in microalgae as a dynamic system shaped by environmental variability. We highlight emerging strategies, including bicarbonate utilization, photobioreactor (PBR) innovation, molecular modeling, and genetic rewiring, that support scalable and specific waste-to-energy conversion. Together, these insights can drive the advancement of microalgal systems tailored for dynamic CO2 conditions.

|
Scooped by
?
July 16, 11:14 AM
|
Indigoidine, a natural bio-pigment with a planar conjugated structure comprising two pyrrolidinone rings linked by a central double bond, exhibits unique chromogenic properties. Growing environmental concerns over synthetic and plant-derived pigments have driven interest in microbial pigments as sustainable alternatives. Despite the potential of nonribosomal peptide synthetases (NRPSs) to optimize indigoidine biosynthesis, mechanistic insights into their catalytic functions remain limited. This review systematically outlines the discovery and evolution of indigoidine, focusing on the structural features of NRPSs and associated biosynthetic pathways. It evaluates current strategies for enhancing production, including host engineering, metabolic flux optimization, genome-wide screening, transcriptional regulation, and scale-up processes. Additionally, the applications of indigoidine in textile, agricultural, and biomedical industries are critically examined. By proposing a novel research framework for microbial pigments synthesis optimization, this review contributes theoretical and technical advancements toward sustainable biopigment manufacturing.

|
Scooped by
?
July 16, 10:32 AM
|
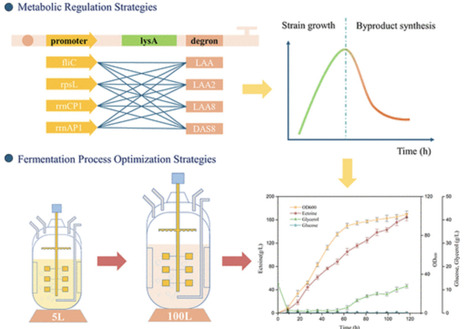
Ectoine is a pivotal natural osmoprotectant that functions as a compatible solute through osmoregulation, enabling microorganisms to thrive in extreme environments such as high salinity. To meet market demands, this study focuses on optimizing its production process. We initially engineered the ectABC gene cluster from Halomonas venusta via 5′-UTR modification, establishing a functional ectoine biosynthesis pathway in E. coli. Subsequent introduction of a rate-limiting enzyme EctB mutant (E407D) and aspartokinase mutant increased titer by 140%. To address lysine byproduct accumulation, an innovative molecular switch was employed to regulate lysA gene expression, achieving dynamic balance between cell growth and product synthesis. Further optimization through cofactor engineering yielded the final strain ECT31, which produced 164.6 g/L ectoine in a 100 L bioreactor within 117 h, the highest reported titer for E. coli-based ectoine production to date. The metabolic engineering strategy presented herein establishes a new pdigm for efficient biosynthesis of amino acid derivatives.

|
Scooped by
?
July 16, 10:02 AM
|
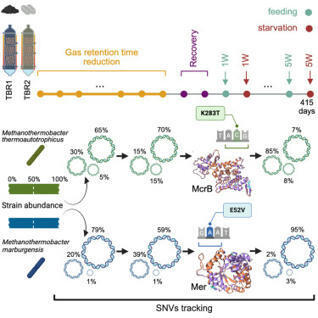
Genetic heterogeneity exists within all microbial populations, with sympatric cells of the same species often exhibiting single-nucleotide variations that influence phenotypic traits, including metabolic efficiency. However, the evolutionary dynamics of these strain-level differences in response to environmental stress remain poorly understood. Here, we present a first-of-its-kind study tracking the adaptive evolution of an anaerobic, carbon-fixing microbiota under a controlled engineered ecosystem focused on carbon dioxide bioconversion into methane. Leveraging strain-resolved metagenomics with an ad hoc variant calling and phasing approach, we mapped mutation trajectories and observed that the two dominant Methanothermobacter species maintained distinct sweeping haplotypes over time, most likely due to niche-specific metabolic roles. By combining population genetic statistics and peptide reconstruction, mer and mcrB genes emerged as potential drivers of archaeal strain-level competition. These findings pave the way for targeted engineering of microbial communities to enhance bioconversion efficiency, with significant implications for sustainable energy and carbon management in anaerobic systems.

|
Scooped by
?
July 16, 9:48 AM
|
Bifidobacteria are beneficial saccharolytic microbes that are widely used as probiotics or in synbiotic formulations, yet individual responses to supplementation can vary with strain type, microbiota composition, diet and lifestyle, underscoring the need for strain-level insights into glycan metabolism. Here we reconstructed 68 pathways for the utilization of mono-, di-, oligo- and polysaccharides by analysing the distribution of 589 curated metabolic gene functions (catabolic enzymes, transporters and transcriptional regulators) across 3,083 non-redundant Bifidobacterium genomes of human origin. Thirty-eight predicted phenotypes were validated in vitro for 30 geographically diverse strains, supporting genomics-based predictions. Our analysis uncovered extensive inter- and intraspecies functional heterogeneity, including a distinct clade within Bifidobacterium longum that metabolizes α-glucans and Bangladeshi isolates carrying unique gene clusters for xyloglucan and human milk oligosaccharide utilization. This large-scale genomic compendium advances our understanding of bifidobacterial carbohydrate metabolism and can inform the rational design of probiotic and synbiotic formulations tailored to strain-specific nutrient preferences. A comprehensive genomic analysis reveals species- and strain-level heterogeneity in carbohydrate utilization potential across bifidobacteria of human origin.
|

|
Scooped by
?
Today, 1:44 AM
|
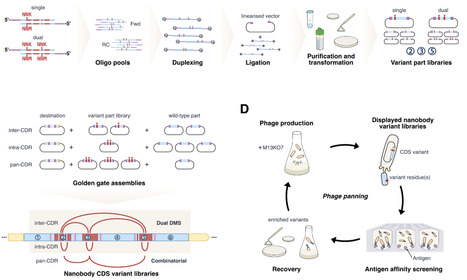
Control over mutational library diversity is an essential consideration when engineering proteins, but is often fraught with trade-offs between diversity, specificity, and affordability. Contemporary library assembly approaches often incorporate oligonucleotide pool synthesis to achieve affordable, precise mutagenesis; however, these oligos are often reliant on complex designs to facilitate downstream PCR and/or restriction digests. Direct hybridization of oligo pools is an overlooked strategy to simplify mutagenesis, especially when paired with a type IIS restriction cloning approach. We validate this approach by designing, hybridising, and deep sequencing single and dual substitution CDR region parts derived from nanobody GA10. Assembly of these parts into a full-length nanobody CDS facilitated the phage display of variant libraries for affinity maturation against its cyclic peptide target. Variants identified through enrichment analysis were expressed in isolation and yielded improved affinities by more than 100-fold. Recent advances in machine learning have successfully inferred improved variants outside of screened library space, but require controlled, multi-mutant libraries. The library assembly approach outlined in this research is well-suited for such approaches.

|
Scooped by
?
Today, 1:16 AM
|
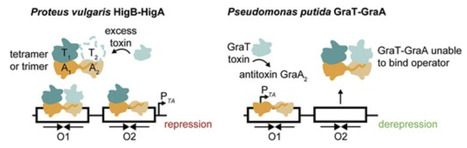
Bacterial toxin–antitoxin (TA) pairs transcriptionally autoregulate their expression via a repression/derepression mechanism in response to changing environmental conditions. The structural diversity of TA systems influences the mechanisms of transcriptional regulation. Here, we define the molecular mechanism for the plasmid-encoded HigB–HigA TA pair originally identified in a post-operative infection with antibiotic-resistant Proteus vulgaris. We determine DNA binding and promoter activity by the HigB–HigA complex supported by structural biology and molecular dynamics simulations of an elusive DNA operator–TA repressor complex. To define the optimal oligomeric TA repressor–DNA operator complex required for derepression, we engineered a dedicated trimeric HigB–HigA2 complex that represses transcription more than 26-fold as compared to the tetrameric HigB2–HigA2. These results expand the known diversity of how the HigB–HigA TA family is autoregulated.

|
Scooped by
?
Today, 12:53 AM
|
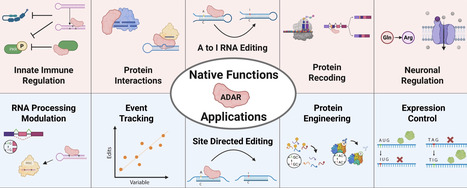
RNA processing is essential for proper cellular function, contributing to protein and cell state diversity, and is often dysregulated in diseased states. A key subset of RNA regulators is the double-stranded RNA-specific adenosine deaminase (ADAR) protein family, which hydrolytically deaminates double-stranded RNA, causing an adenosine-to-inosine edit (A-to-I). Active ubiquitously throughout the body, this pleiotropic protein family plays critical roles in embryonic patterning, neurological function, and immune regulation. Their aberrant activity has in turn been implicated in a spectrum of disorders, including cancer, metabolic diseases, and autoimmune conditions. By instead purposefully modulating their activity, ADARs have been leveraged to create a versatile toolset for transcriptome engineering. This includes enabling programmable RNA editing, controlled RNA splicing, reversibly modulating protein interactions, and altering cellular inflammation. Here, we review the pleiotropic functions and versatile applications of ADARs, as well as outline areas for growth and potential new avenues in both therapeutics and research.

|
Scooped by
?
Today, 12:49 AM
|
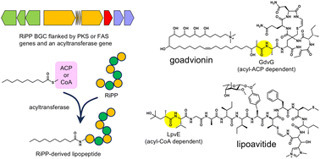
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a diverse superfamily of natural products unified by a common biosynthetic logic: The peptide backbone is genetically encoded, and the translated precursor peptide undergoes a series of post-translational modifications catalyzed by maturase enzymes to produce the final bioactive compound. Despite their structural complexity, RiPPs are encoded by relatively small biosynthesis gene clusters. RiPP maturase enzymes are diverse and often promiscuous, offering significant biotechnological potential. However, their lack of conserved features makes genome-based discovery of novel RiPPs challenging. Recent advances in biosynthetic understanding and genome mining techniques have led to the identification of numerous uncharacterized RiPP biosynthetic gene clusters, often flanked by genes encoding non-RiPP moieties, in microbial genomes. Leveraging this information, a new class of natural products, hybrids of RiPPs and non-RiPP elements, has recently been discovered. Among them, RiPPs bearing fatty acyl groups, referred to as RiPP-derived lipopeptides, represent a newly emerging class of lipopeptide natural products with significant antimicrobial activity. bgc

|
Scooped by
?
Today, 12:35 AM
|
Whole-cell biosensors are powerful tools for metabolite monitoring, yet challenges such as narrow dynamic range and high leaky expression limit their broader applications. Here, we present a systematic workflow based on two Design-Build-Test-Learn (DBTL) cycles to develop and optimize a transcription factor-based pyruvate biosensor in Escherichia coli. In the first iteration of the cycle, we constructed a biosensor that responded to intracellular pyruvate levels within 0.05 - 10mM range. In the second cycle, we implemented design of experiment (DoE) to systematically explore combinatorial effects of promoters and ribosome binding sites (RBSs). A first set of experiments were designed to identify factors with a significant effect on biosensor performance. The results showed RBS of report gene significantly influenced dynamic range by modulating basal and maximum expression, while RBS of transcription factor affected signal span. The Akaike Information Criterion was used to select a model incorporating two main effects and one interaction effects. The best-performing strain exhibited an 18.5-fold increase in dynamic range and a 37.2-fold reduction in leaky expression. Quantification of intracellular pyruvate confirmed an operational range of 1.23 - 6.81 μmol/g DCW. Our work demonstrates the power of DBTL cycles with statistical modelling for biosensor engineering, enabling more precise metabolic regulation and screening applications. sensor

|
Scooped by
?
July 16, 11:10 PM
|
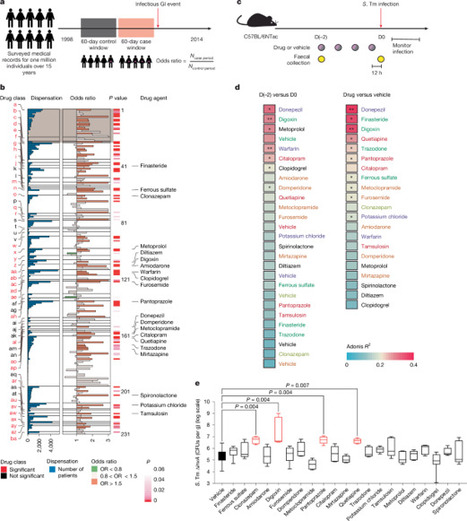
Most people in the USA manage their health by taking at least one prescription drug, and drugs classified as non-antibiotics can adversely affect the gut microbiome and disrupt intestinal homeostasis. Here we identify medications that are associated with an increased risk of gastrointestinal infections across a population cohort of more than one million individuals monitored over 15 years. Notably, the cardiac glycoside digoxin and other drugs identified in this epidemiological study are sufficient to alter the composition of the microbiome and the risk of infection with Salmonella enterica subsp. enterica serovar Typhimurium (S. Tm) in mice. The effect of digoxin treatment on S. Tm infection is transmissible through the microbiome, and characterization of this interaction highlights a digoxin-responsive β-defensin that alters the microbiome composition and consequent immune surveillance of the invading pathogen. Combining epidemiological and experimental approaches thus provides an opportunity to uncover drug–host–microbiome–pathogen interactions that increase the risk of infections in humans. An analysis of prescription medications shows that several non-antibiotic drugs, such as the heart medication digoxin, can reduce the immune response to pathogens and increase the risk of gastrointestinal infections by altering the composition of the microbiome.

|
Scooped by
?
July 16, 10:56 PM
|
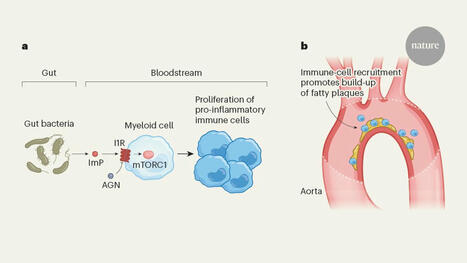
a, Mastrangelo et al. show that the molecule imidazole propionate (ImP), which is made by bacteria in the gut, enters the bloodstream and interacts with the imidazoline 1 receptor (I1R). I1R is expressed in a class of blood cell known as myeloid cells. Activation of I1R stimulates the protein complex mTORC1, which leads to the proliferation of pro-inflammatory immune cells. b, This results in immune cells being recruited to fatty plaques on the walls of blood vessels such as the aorta, the body’s largest artery. The build-up of cholesterol and immune cells is known as atherosclerosis, and can result in obstruction of blood vessels and further cardiovascular problems. Inhibiting I1R using a compound called AGN (shown in a) reverses this pro-atherosclerotic effect.

|
Scooped by
?
July 16, 10:23 PM
|
Remodelling plant immune receptors has become a vital strategy for creating new disease resistance traits to combat the growing threat of plant pathogens to global food security and environmental sustainability. However, current methods are constrained by the rapid evolution of plant pathogens and often lack broad-spectrum and durable protection. Here we report an innovative strategy to engineer broad-spectrum, durable and complete disease resistance in plants through expression of a chimeric protein containing a flexible polypeptide coupled with a single or dual conserved pathogen-originated protease cleavage site fused in frame to the N terminus of an autoactive nucleotide-binding and leucine-rich-repeat immune receptor (NLR) containing a coiled-coil or RESISTANCE TO POWDERY MILDEW 8-like coiled-coil domain. Following invasion, pathogen-originated specific proteases cleave the inactive chimeric protein to form free autoactive NLR, triggering broad-spectrum plant disease resistance. We demonstrate that a single engineered NLR can confer broad-spectrum and complete resistance against multiple potyviruses. Given that many pathogenic organisms across kingdoms encode proteases, this strategy has the potential to be exploited to control viruses, bacteria, oomycetes, fungi, nematodes and pests in plants. Cleavage by pathogen-derived proteases of an engineered chimeric protein activates its plant immune receptor component, enabling broad-spectrum resistance to pathogens in plants.

|
Scooped by
?
July 16, 11:19 AM
|
Gene mutation is the primary source of genetic variation, yet natural mutation rates are insufficient to meet the demands of biotechnological applications. This review systematically examines advancements in hypermutation technologies designed to overcome this limitation, focusing on targeted, multi-target, and genome-wide approaches, and their transformative applications across diverse fields. By comparing these tools across multiple dimensions, including mutation scope, rate, and type, we provide insights to guide the selection of optimal mutagenesis tools for accessing distinct genetic landscapes and addressing practical challenges. Furthermore, we summarize the challenges that persist in this active area and highlight future directions such as developing high-throughput screening methods and AI-driven predictive models for mutational outcomes. By bridging the gap between natural mutation constraints and biotechnological needs, hypermutation technologies promise to unlock unprecedented innovations in synthetic biology, evolutionary research, and industrial applications and pave the way for exploring previously inaccessible genetic landscapes.

|
Scooped by
?
July 16, 11:06 AM
|
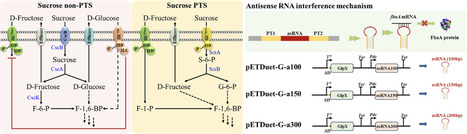
d-Allulose, a rare sugar characterized by its high sweetness and low-calorie profile, is gaining attention in the sweetener market. This study introduces an innovative method for converting sucrose into d-allulose through microbial fermentation. An irreversible synthesis pathway was constructed by expressing the scrA, scrB, alsE, and a6PP genes in Escherichia coli JM109 (DE3), enhancing substrate utilization via dual PTS-dependent transport of sucrose and d-fructose. A fructose-1,6-bisphosphatase mutant (GlpX [K29A]) was used to facilitate the influx of fructose-1-phosphate into the synthesis pathway. The Embden–Meyerhof–Parnas (EMP) and pentose phosphate (PP) pathways were weakened by deleting the pfkA and rpiA genes. To further regulate carbon fluxes, a structurally stable antisense RNA (asRNA) was employed to inhibit FbaA expression. The fermentation medium was optimized using response surface methodology. Finally, the d-allulose titer reached 12.8 g/L, with a yield of 0.23 g/g on sucrose, achieved through fed-batch fermentation in a 5 L fermenter.

|
Scooped by
?
July 16, 10:23 AM
|
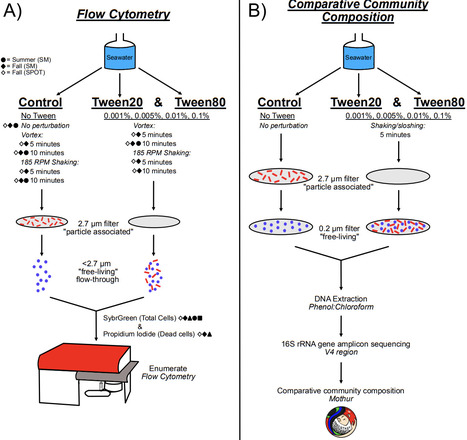
Marine particles, typically composed of organic detritus and cellular debris, harbor microbial communities that are distinct from the planktonic, or free-living, communities in the pelagic ocean. However, without being first separated from the particle and microbial consortia, these microbes are inaccessible to further investigation via single-cell microbiology methods like flow cytometry, cell-sorting, and dilution-based isolation. To confront this obstacle, we compared the dissociative effects of two commonly used detergents, Tween20 and Tween80, on particle-associated marine microbial communities. The ability of Tween treatments to liberate cells from particles, and to maintain cell integrity, was quantified by flow cytometry from multiple communities across seasons and locations. Both Tween20 and Tween80, at 185 RPM shaking, gently dissociated microbes from their particles, causing very little cell mortality. Additionally, Tween80 liberated a greater number of particle-associated cells into the free-living fraction. We also analyzed the effects of Tween treatments on the microbial community composition for one of these collections via 16S rRNA gene amplicon sequencing of the particle-associated and free-living fractions relative to unamended controls. Tween20 and Tween80 were both effective for microbial dissolution from particles, however Tween80 treatments displayed greater uniformity in the dissociated communities and significantly enriched for the most abundant particle-associated members. Together these data indicate that Tween80 was most effective at gently dissociating particle-associated cells.

|
Scooped by
?
July 16, 9:51 AM
|
Plasmids constitute a key tool in synthetic biology, providing a versatile framework for various research and industrial ventures. An essential determinant of plasmid functionality is its copy number, impacting both protein production rates and host cell metabolic burden. However, currently there is no model that can computationally predict the plasmid copy number from the sequence of its origin of replication (ORI). We present a novel software solution tailored to simplify plasmid copy number design, poised to redefine plasmid engineering workflows. At the heart of our tool lies a comprehensive machine learning model, informed by numerous features extracted from the ORI. This computational model emphasizes the importance of promoter strength and the RNA folding dynamics of regulatory elements within the ORI. Additionally, we detail a robust protocol for the efficient manipulation of plasmid ORIs used to validate our model’s predictive capabilities. This innovation represents a paradigm shift in plasmid-centric methodologies, offering unprecedented avenues for advancement in synthetic biology research and industrial applications. The software is available at: https://pcn-gradient.vercel.app/
|
 Your new post is loading...
Your new post is loading...