Your new post is loading...

|
Scooped by
?
Today, 1:31 AM
|
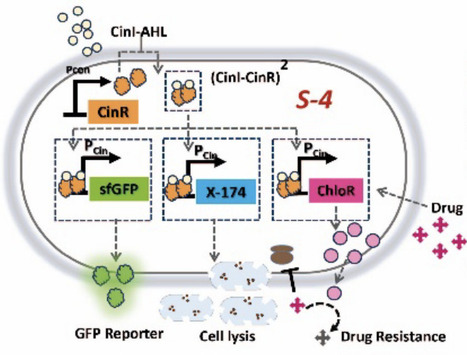
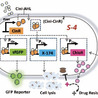
Synthetic biology enables the precise control of bacterial population dynamics through engineered genetic circuits, paving the way for innovative therapeutic and biosensing applications. Here, we present genetic circuit design that leverages AHL (acyl-homoserine lactone)-based quorum sensing and a transcriptional cascade regulated by an external inducer to drive synchronized proliferation and lysis events. In this system, the absence of the inducer arabinose prevents CinR mediated repression, enabling activation of a transcriptional cascade that drives the expression of the lysis protein, fluorescent protein, and a drug resistance gene. The accumulation of AHL acts as a proxy for population density and modulates gene expression through a quorum-sensing mechanism. This inducible circuit provides tunable and pulsatile control over bacterial population dynamics, enabling cycles of population growth and collapse. Such behavior prevents premature population clearance while ensuring functional bacterial densities for extended durations. Our system represents a foundational step toward implementing more complex multi-stable architectures for self-regulation of bacterial population. This dynamic and modular framework offers significant advantages for therapeutic interventions, such as self-regulating drug delivery, inflammation-responsive probiotic therapies, and targeted bacterial therapy.

|
Scooped by
?
Today, 1:13 AM
|
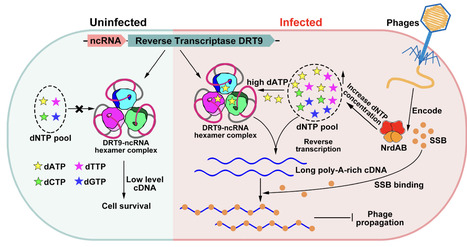
Prokaryotic defense-associated reverse transcriptases (DRTs) were recently identified with antiviral functions; however, their functional mechanisms remain largely unexplored. Here we show that DRT9 forms a hexameric complex with its upstream non-coding RNA (ncRNA) to mediate antiphage defense by inducing cell growth arrest via abortive infection. Upon phage infection, the phage-encoded ribonucleotide reductase NrdAB complex elevates intracellular dATP levels, activating DRT9 to synthesize long, poly-A-rich single-stranded cDNA, which likely sequesters the essential phage SSB protein and disrupts phage propagation. We further determined the cryo-electron microscopy structure of the DRT9-ncRNA hexamer complex, providing mechanistic insights into its cDNA synthesis. These findings highlight the diversity of RT-based antiviral defense mechanisms, expand our understanding of RT biological functions, and provide a structural basis for developing DRT9-based biotechnological tools.

|
Scooped by
?
Today, 12:02 AM
|
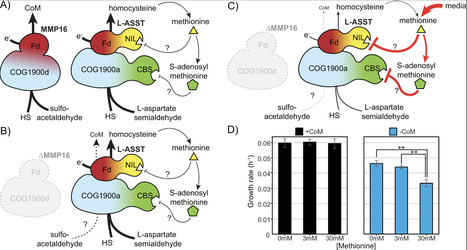
2-mercaptoethanesulfonate (Coenzyme M, CoM) is an organic sulfur-containing cofactor used for hydrocarbon metabolism in archaea and bacteria. In archaea, CoM serves as an alkyl group carrier for enzymes belonging to the alkyl-CoM reductase family, including methyl-CoM reductase, which catalyzes methane formation in methanogens. Two pathways for the biosynthesis of CoM have been identified in methanogenic archaea. The initial steps of these pathways are distinct but the last two reactions, leading up to CoM formation, are universally conserved. The final step is proposed to be mediated by methanogenesis marker metalloprotein 16 (MMP16), a putative sulfurtransferase, that replaces the aldehyde group of sulfoacetaldehyde with a thiol to generate CoM. Based on prior research, assignment of MMP16 as CoM synthase (ComF) is not widely accepted as deletion mutants have been shown to grow without any CoM dependence. Here, we investigate the role of MMP16 in the model methanogen, Methanosarcina acetivorans. We show that a mutant lacking MMP16 has a CoM-dependent growth phenotype and a global transcriptomic profile reflective of CoM-starvation. Additionally, the ∆ MMP16 mutant is a CoM auxotroph in sulfide-free medium. These data reinforce prior claims that MMP16 is a bona fide ComF but point to backup pathway(s) that can conditionally compensate for its absence. We found that L-aspartate semialdehyde sulfurtransferase (L-ASST), catalyzing a sulfurtransferase reaction during homocysteine biosynthesis in methanogens, is potentially involved in genetic compensation of the MMP16 deletion. Even though both L-ASST and MMP16 are members of the COG1900 family, site-directed mutagenesis of conserved cysteine residues implicated in catalysis reveal that the underlying reaction mechanisms may be distinct.

|
Scooped by
?
May 4, 11:54 PM
|
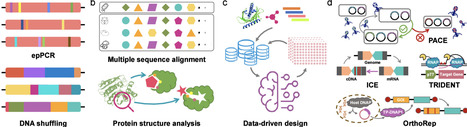
The booming field of synthetic biology and metabolic engineering provides promising approaches for sustainable manufacturing of chemicals from renewable feedstocks with microbial cell factories. Classical metabolic engineering strategies mainly focus on altering gene expression levels and enzyme concentrations to improve the metabolic fluxes of specific pathways. However, the impact and limitations of enzyme properties, which are usually ignored in classical metabolic engineering efforts, can hinder further optimization of microbial cell factories. Protein engineering and directed evolution are powerful tools for modifying proteins to achieve desirable properties, and they have been integrated into metabolic engineering efforts to build highly efficient metabolic pathways and optimal industrial chassis. In this review, we present traditional and data-driven strategies and techniques of directed evolution, including random library design, semirational design, smart library design, and in vivo continuous evolution. We also discuss how these directed evolution strategies have been applied in metabolic engineering toward superphenotypes that cannot be achieved through simple gene overexpression or knockout. Finally, we discuss the challenges of applying protein engineering in metabolic engineering and the prospects for accelerating the directed evolution workflow using the state-of-art technologies.

|
Scooped by
?
May 4, 11:43 PM
|
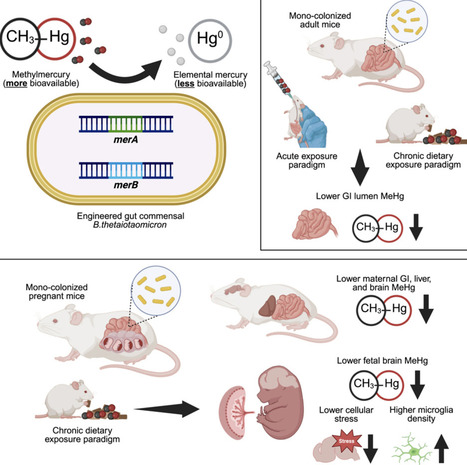
Despite efforts to decrease mercury emissions, chronic exposure to the neurotoxicant methylmercury (MeHg) continues to be a global problem that contributes to disparities in risk for neurological and metabolic diseases. Herein we engineer a human commensal gut bacterium, Bacteroides thetaiotaomicron (Bt), to detoxify MeHg by heterologous expression of organomercury lyase (MerB) and mercuric reductase (MerA) genes derived from a resistant bacterium isolated from Hg-polluted mines. We demonstrate that BtmerA/B demethylates MeHg both in vitro and within the intestines of mice orally exposed to MeHg or diets containing MeHg-rich fish. In pregnant mice exposed to dietary MeHg, BtmerA/B decreases MeHg accumulation in the maternal liver, brain, placenta, and fetal brain, and attenuates the expression of cellular stress genes in the fetal brain. Overall, this work provides foundational proof-of-principle supporting the ability of an engineered gut bacterium to limit MeHg bioaccumulation and reduce adverse effects of chronic MeHg exposure.

|
Scooped by
?
May 4, 11:12 PM
|
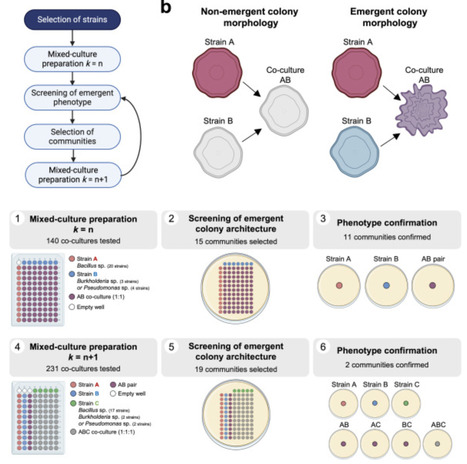
Synthetic communities (SynComs) are valuable tools for addressing microbial community assembly and function, towards their manipulation for clinical, biotechnological and agricultural applications. However, SynCom design is complicated since interactions between microbes cannot be predicted based on their individual properties. Here we aimed to assemble a functionally cohesive SynCom displaying high-order interactions, as a model to study the community-level beneficial functions of seed-endophytic bacteria from native maize landraces, including strains from the Bacilli class, and the Burkholderia and Pseudomonas genera. We developed a partial combinatorial, bottom-up strategy that was followed by the detection of complex colony architecture as an emergent property in co-cultures. Using this simplified approach, we tested less than 400 co-cultures from a pool of 27 strains, resulting in the assembly the Xilonen SynCom, which includes Bacillus pumilus NME155, Burkholderia contaminans XM7 and Pseudomonas sp. GW6. In this community, higher-order interactions result in complex colony architecture, which is considered a proxy of biofilm formation. Additionally, we generated protocols for absolute quantification of each member from a complex mixture. The Xilonen SynCom will serve as a model to study biofilm formation in community settings, and will aid in the study of the molecular and ecological basis mediating maize fertility.

|
Scooped by
?
May 4, 10:59 PM
|
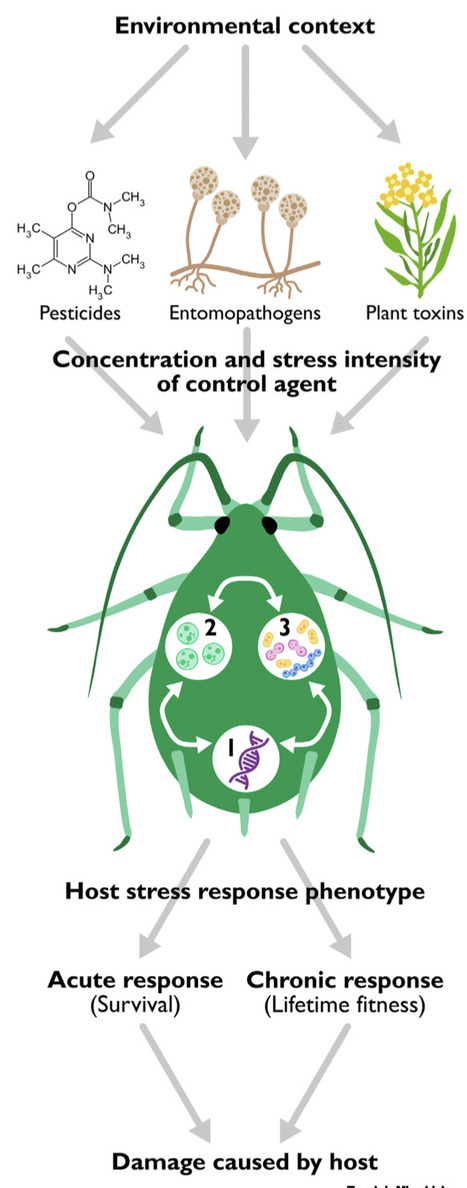
Bacterial symbionts in pests are being increasingly investigated to assess their potential uses for sustainable control approaches. We undertook a review and analysis of the impacts of endosymbionts and gut symbionts on responses to toxins from plants and pesticides, and to attack by fungal entomopathogens. Despite methodological issues affecting estimates of effect sizes, there is evidence for symbionts increasing resistance to all three agents. However, impacts can be small, and for pesticides, these may not reach levels required for resistance at field rates. Negative or neutral effects may be underreported. Further complications arise because host genotype and the environment impact symbiont effects. We anticipate rapid progress in this area over coming years that should clarify practical implications of these effects.

|
Scooped by
?
May 4, 10:48 PM
|
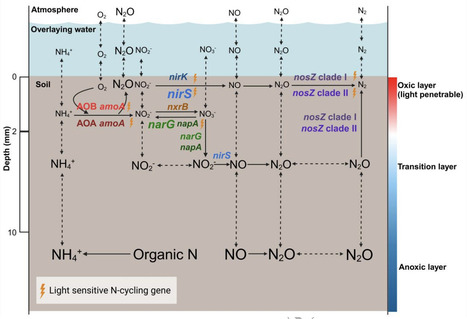
Wetlands can be a significant source of N2O under current global climate change regime with the soil-water interface representing a biogeochemical hotspot for microbial activity. However, the role of soil-water interface in controlling N2O emissions remains poorly understood. We hypothesized that the millimeter-scale redox gradient across the soil-water interface generates corresponding distinct niche for N-cycling microorganisms that collectively regulate the production and consumption of N2O over the same spatial scale. The abundance, transcriptional activity and spatial organization of different N-cycling guilds across the soil-water interface were characterized in mesocosms from three different paddy soils with different N2O emissions. Results demonstrated millimeter-scale stratification of N-cycling microbial activity across the soil-water interface, and in particular within the first 10 mm of flooded soils. Ammonia-oxidizing microorganisms were only transcriptionally active in the top 4 mm, suggesting a previously underestimated contribution to N2O emissions from wetlands. Variation in N2O accumulation was observed across the soil-water interface, with the highest concentrations measured at either the soil-water interface or in the deeper anoxic layer of paddy soils. Despite this difference, N2O-reducing microorganisms exhibited high transcriptional activity at the soil-water interface in all soils, suggesting that there is a microbial-mediated sink for N2O across the soil-water interface that can reduce N2O produced from both oxic and anoxic layers. This work demonstrate an underappreciated and essential role of the microbial hot zones at soil-water interface in regulating N2O emissions from wetlands.

|
Scooped by
?
May 4, 9:50 PM
|
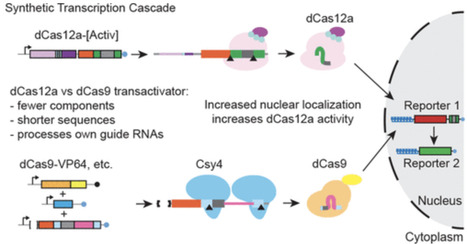
Gene regulatory networks, which control gene expression patterns in development and in response to stimuli, use regulatory logic modules to coordinate inputs and outputs. One example of a regulatory logic module is the gene regulatory cascade (GRC), where a series of transcription factor genes turn on in order. Synthetic biologists have derived artificial systems that encode regulatory rules, including GRCs. Furthermore, the development of single-cell approaches has enabled the discovery of gene regulatory modules in a variety of experimental settings. However, the tools available for validating these observations remain limited. Based on a synthetic GRC using DNA cutting-defective Cas9 (dCas9), we designed and implemented an alternative synthetic GRC utilizing DNA cutting-defective Cas12a (dCas12a). Comparing the ability of these two systems to express a fluorescent reporter, the dCas9 system was initially more active, while the dCas12a system was more streamlined. Investigating the influence of individual components of the systems identified nuclear localization as a major driver of differences in activity. Improving nuclear localization for the dCas12a system resulted in 1.5-fold more reporter-positive cells and a 15-fold increase in reporter intensity relative to the dCas9 system. We call this optimized system the “Synthetic Gene Regulatory Network” (SGRN, pronounced “sojourn”).

|
Scooped by
?
May 4, 9:27 PM
|
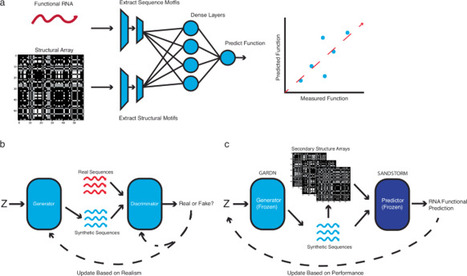
RNA is a remarkably versatile molecule that has been engineered for applications in therapeutics, diagnostics, and in vivo information-processing systems. However, the complex relationship between the sequence, structure, and function of RNA often necessitates extensive experimental screening of candidate sequences. Here we present a generalized, efficient neural network architecture that utilizes the sequence and structure of RNA molecules (SANDSTORM) to inform functional predictions across a diverse range of settings. We pair these predictive models with generative adversarial RNA design networks (GARDN), allowing the generative modelling of a diverse range of functional RNA molecules with targeted experimental attributes. This approach enables the design of novel sequence candidates that outperform those encountered during training or returned by classical thermodynamic algorithms, and can be deployed using as few as 384 example sequences. SANDSTORM and GARDN thus represent powerful new predictive and generative tools for the development of RNA molecules with improved function. Designing high performance RNA molecules is a unifying challenge across many areas of biotechnology research. Here, authors develop GARDN and SANDSTORM, a data-efficient generative AI framework for designing a diverse set of RNA molecules. These tools represent a promising addition to the sequence design toolkit that could be used in many domains.

|
Scooped by
?
May 4, 5:19 PM
|
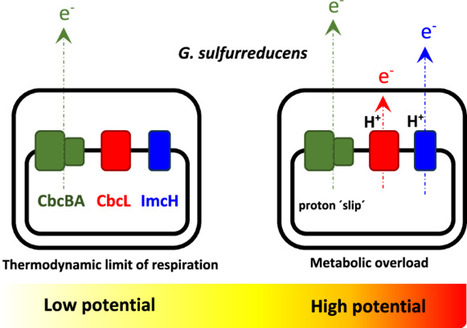
Studies on electron-transfer pathways in certain bacterial strains have revealed that the degree of coupling of electron transfer to proton translocation along the respiratory chain can be regulated according to metabolic demands. This first line of metabolic response, based on the existence of energy dissipation mechanisms, has not been demonstrated to be a general pattern across the bacterial kingdom, let alone to be operative in electro-active bacteria. In this study, we hypothesized that electro-active cells should respond to over-polarization by also triggering energy decoupling mechanisms to prevent metabolic overloads. Based on electrochemical analyses, we propose that the recently discovered inner-membrane cytochrome CbcBA - used by electro-active Geobacter sulfurreducens bacteria for cellular respiration near the thermodynamic energetic limit - can also act as an energy dissipation gate when the metabolism is demanded, contributing to regulate the energy balance of the cell by decoupling carbon assimilation from electrode respiration. In response to extreme electrode potentials, electroactive Geobacter sulfurreducens uses the inner-membrane cytochrome CbcBA as an energy dissipation gate, regulating the energy balance of the cell by decoupling carbon assimilation from electrode respiration.

|
Scooped by
?
May 4, 4:59 PM
|

|
Scooped by
?
May 4, 4:54 PM
|
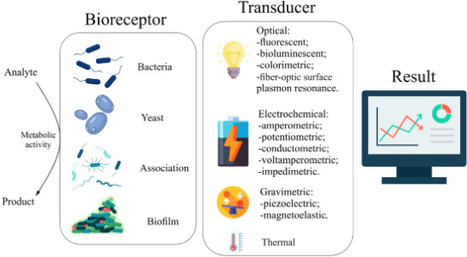
Microbial biosensors are bioanalytical devices that can measure the toxicity of pollutants or detect specific substances. This is the greatest advantage of microbial biosensors which use whole cells of microorganisms as powerful tools for measuring integral parameters of environmental pollution. This review explores the core principles of microbial biosensors including biofuel devices, emphasizing their capacity to evaluate biochemical oxygen demand (BOD), toxicity, heavy metals, surfactants, phenols, pesticides, inorganic pollutants, and microbiological contamination. However, practical challenges, such as sensitivity to environmental factors like pH, salinity, and the presence of competing substances, continue to hinder their broader application and long-term stability. The performance of these biosensors is closely tied to both technological advancement and the scientific understanding of biological systems, which influence data interpretation and device optimization. The review further examines cutting-edge developments, including the integration of electroactive biofilms with nanomaterials, molecular biology techniques, and artificial intelligence, all of which significantly enhance biosensor functionality and analytical accuracy. Commercial implementations and improvement strategies are also discussed, providing a comprehensive overview of the state-of-the-art in this field. Overall, this work consolidates recent progress and identifies both the potential and limitations of microbial biosensors, offering valuable insights into their future development for environmental monitoring.
|

|
Scooped by
?
Today, 1:24 AM
|
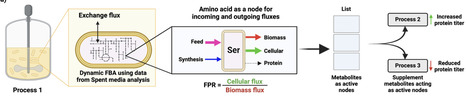
Complex media supplemented with a carbon source are commonly used in bioprocesses for recombinant protein production in Escherichia coli. Optimizing these processes is challenging and requires precise understanding of cellular metabolism and nutrient requirements. Compared to a design of experiments approach that necessitates extensive experimentation, metabolic modeling using a genome scale metabolic model (GEM) offers a more predictive and systematic approach to guide process optimization by identifying specific metabolic bottlenecks. In addition, spent media analysis (SMA) can unravel the preferential utilization of different media components during the bioprocess. Here, we integrated the updated E. coli GEM with time course SMA data from a fed-batch process and performed dynamic flux balance analysis (dFBA) to identify metabolites that function as active nodes and are vital for cellular function. These are potential target supplements to boost cellular activity and in turn the recombinant protein productivity. Using an iterative approach of performing fermentation, SMA, and metabolic modeling, we intensified the bioprocess in just five experimental trials, resulting in a six-fold increase in protein productivity. Our new feeding strategy involved yeast extract with amino acid supplementation (Ser, Thr, Asp, and Glu) and increased oxygen transfer rates. This approach demonstrates significant promise for application in bioprocess intensification.

|
Scooped by
?
Today, 1:09 AM
|
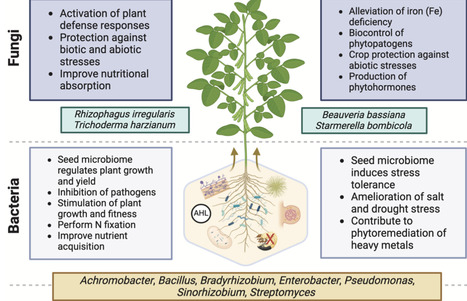
In the context of global challenges such as climate change, soil degradation, and food security, understanding the modes of action of Plant Growth-Promoting Microorganisms (PGPMs), their formulation, and their application is crucial and can be more focused in future research projects. This editorial paper aims to elucidate diverse modes of action employed by different types of PGPMs, including nitrogen fixation, phosphorus solubilization, production or regulation of phytohormones, and plant protection against environmental and biotic stresses as demonstrated and discussed in the Special Issue entitled “Plant Growth-Promoting Microorganisms: new insights and the way forward”.

|
Scooped by
?
May 4, 11:58 PM
|
Aspergillus fumigatus is a saprophytic fungus prevalent in the environment and capable of causing severe invasive infection in humans. This organism can use strategies such as molecule masking, immune response manipulation and gene expression alteration to evade host defences. Understanding these mechanisms is essential for developing effective diagnostics and therapies to improve patient outcomes in Aspergillus-related diseases. In this Review, we explore the biology and pathogenesis of A. fumigatus in the context of host biology and disease, highlighting virus-associated pulmonary aspergillosis, a newly identified condition that arises in patients with severe pulmonary viral infections. In the post-pandemic landscape, in which immunotherapy is gaining attention for managing severe infections, we examine the host immune responses that are critical for controlling invasive aspergillosis and how A. fumigatus circumvents these defences. Additionally, we address the emerging issue of azole resistance in A. fumigatus, emphasizing the urgent need for greater understanding in an era marked by increasing antimicrobial resistance. This Review provides timely insights necessary for developing new immunotherapeutic strategies against invasive aspergillosis. In this Review, van de Veerdonk and colleagues explore Aspergillus fumigatus biology, the host immune responses and the fungal mechanisms of immune evasion, as well as emerging challenges such as virus-associated pulmonary aspergillosis and azole resistance, and reflect on future directions for therapeutics and patient care improvement.

|
Scooped by
?
May 4, 11:47 PM
|
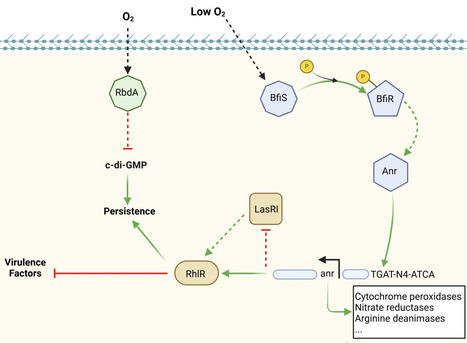
The exquisite ability of bacteria to adapt to their environment is essential for their capacity to colonize hostile niches. In the cystic fibrosis (CF) lung, hypoxia is among several environmental stresses that opportunistic pathogens must overcome to persist and chronically colonise. Although the role of hypoxia in the host has been widely reviewed, the impact of hypoxia on bacterial pathogens has not yet been studied extensively. This review considers the bacterial oxygen-sensing mechanisms in three species that effectively colonise the lungs of people with CF, namely Pseudomonas aeruginosa, Burkholderia cepacia complex and Mycobacterium abscessus and draws parallels between their three proposed oxygen-sensing two-component systems: BfiSR, FixLJ, and DosRS, respectively. Moreover, each species expresses regulons that respond to hypoxia: Anr, Lxa, and DosR, and encode multiple proteins that share similar homologies and function. Many adaptations that these pathogens undergo during chronic infection, including antibiotic resistance, protease expression, or changes in motility, have parallels in the responses of the respective species to hypoxia. It is likely that exposure to hypoxia in their environmental habitats predispose these pathogens to colonization of hypoxic niches, arming them with mechanisms than enable their evasion of the immune system and establish chronic infections. Overcoming hypoxia presents a new target for therapeutic options against chronic lung infections.

|
Scooped by
?
May 4, 11:42 PM
|
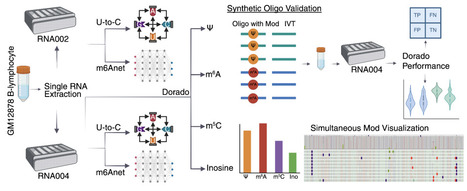
Nanopore technology can directly sequence intact RNA molecules, offering a unique capability to read native modifications. Oxford Nanopore Technologies recently updated its direct RNA sequencing technology from RNA002 to RNA004 chemistry. This update included an improved basecaller (Dorado) for increased sequencing accuracy, and ionic current models for de novo detection of four RNA modifications. Using a single RNA extraction from GM12878 B-lymphocyte cell line, we compared RNA002 and RNA004 sequencing chemistries and evaluated Dorado modification calling accuracy. We computed U-to-C mismatches, previously used to identify putative pseudouridine sites, and ran m6anet for identifying putative N6-methyladenosine sites. Dorado results for each respective modification showed both global and site-specific differences when compared to RNA002 results. We used DRS data from in vitro transcription of GM12878 genomic DNA as well as synthetic oligonucleotides to evaluate Dorado modification calling performance. Dorado pseudouridine model achieved 96-98% for both accuracy and F1-score. Similarly, Dorado N6-methyladenosine model achieved 94-98% accuracy, 96-99% F1-score. Our results demonstrate that Nanopore Direct RNA sequencing could simultaneously detect pseudouridine, N6-methyladenosine, 5-methylcytosine, and inosine modifications on individual mRNA strands. It is critical to validate these calls using orthogonal methods (e.g., Liquid Chromatography with Tandem Mass Spectrometry) for increased confidence.

|
Scooped by
?
May 4, 11:05 PM
|
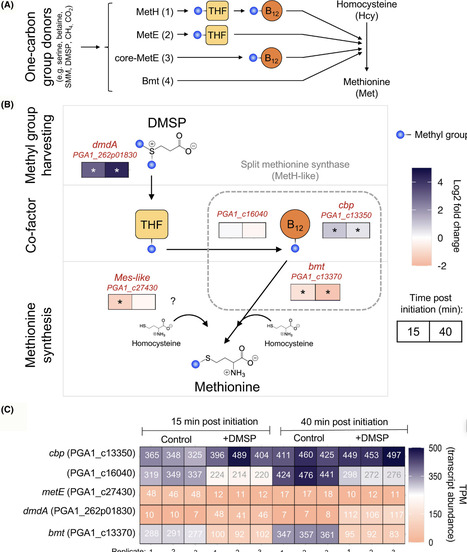
Bacteria can shorten their lag phase by using methyl groups from compounds like dimethylsulfoniopropionate (DMSP), which are incorporated into cellular components via the methionine cycle. However, the role of specific methionine synthases in this process is not fully understood. Using transcriptomics, genetics, and biochemical assays, we investigated methionine synthases involved in lag phase shortening in Phaeobacter inhibens. We focused on a cobalamin-dependent methionine synthase (MetH)-like complex encoded by three genes: a betaine-homocysteine S-methyltransferase (bmt), a cobalamin-binding protein (cbp), and an intermediate methyl carrier (PGA1_c16040). Expression profiling revealed transcriptional decoupling among these genes. Deleting bmt disrupted lag phase shortening in response to DMSP. Functional assays showed that Bmt can directly produce methionine from DMSP and betaine, independent of tetrahydrofolate (THF) or cobalamin. Interestingly, under stress conditions, lag phase shortening occurred even in the absence of dimethylsulfoniopropionate demethylase DmdA, the primary DMSP demethylase. Under osmotic and oxidative stress, bmt expression increased significantly in response to both DMSP and betaine, suggesting an alternative methylation route. This highlights the role of Bmt as both demethylase and a methionine synthase under stress, offering a cost-effective strategy for methyl group assimilation. Our findings reveal a novel stress-responsive pathway for methionine synthesis and demonstrate the role of Bmt in promoting bacterial adaptation by accelerating the lag phase.

|
Scooped by
?
May 4, 10:52 PM
|
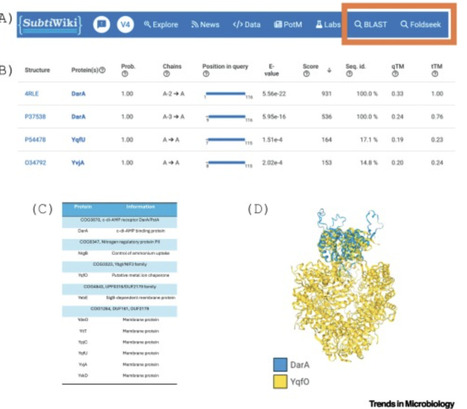
The recent application of artificial intelligence-based prediction of protein and protein complex structures by AlphaFold has revolutionized biology. Here, we give an overview of the tools that make this revolution accessible to experimental microbiologists without training in bioinformatics or sophisticated computer equipment, and show how this helps to gain new insights.

|
Scooped by
?
May 4, 10:40 PM
|
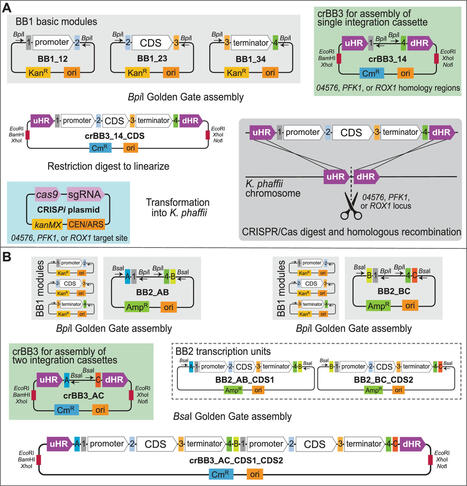
The yeast Komagataella phaffii (formerly known as Pichia pastoris) has been widely used for functional expression of recombinant proteins, including plant and animal food proteins. CRISPR/Cas9 genome editing systems can be used for insertion of heterologous genes without the use of selection markers. The study aimed to create a convenient markerless knock-in method for integrating expression cassettes into the chromosome of K. phaffii using CRISPR/Cas9 technology. The approach was based on the hierarchical, modular, Golden Gate assembly employing the GoldenPiCS toolkit. Furthermore, the aim was to evaluate the system’s efficiency and suitability for producing secreted recombinant food proteins. Three Cas9/sgRNA plasmids were constructed, along with corresponding donor helper plasmids containing homology regions for chromosomal integration via homology-directed repair. The integration efficiency of an enhanced green fluorescent protein (eGFP) expression cassette was assessed at three genomic loci (04576, PFK1, and ROX1). The 04576 locus showed the highest integration efficiency, while ROX1 had the highest transformation efficiency. Whole genome sequencing revealed variable copy numbers of eGFP expression cassettes among clones, corresponding with increasing levels of fluorescence. Furthermore, the system’s applicability for producing recombinant food proteins was validated by successfully expressing and secreting chicken ovalbumin. This constitutes the first report of CRISPR/Cas9 applied to produce recombinant chicken ovalbumin. Conclusions The adapted GoldenPiCS toolkit combined with CRISPR/Cas9 technology enabled efficient and precise genome integration in K. phaffii. This approach holds promise for expanding the production of high-value recombinant proteins. Future research should focus on optimizing integration sites and improving cloning procedures to enhance the system’s efficiency and versatility.

|
Scooped by
?
May 4, 9:43 PM
|
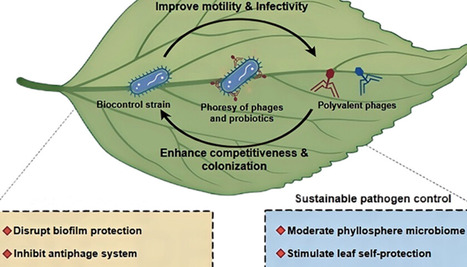
Phage-based biocontrol has shown notable advantages in protecting plants against pathogenic bacteria in agricultural settings compared to chemical-based bactericides. However, the efficiency and scope of phage biocontrol of pathogenic bacteria are limited by the intrinsic properties of phages. Here, we investigated pathogen biofilm eradication in the phyllosphere using the phoresy system of hitchhiking phages onto carrier biocontrol bacteria. The phoresy system efficiently removed the pathogen biofilm in the soybean phyllosphere, reducing the total biomass by 58% and phytopathogens by 82% compared to the untreated control. Biofilm eradication tests demonstrated a significant combined beneficial effect (Bliss independence model, CI < 1) as phages improved carrier bacteria colonization by 1.2-fold and carrier bacteria facilitated phage infection by 1.4-fold. Transcriptomic analysis showed that phoresy significantly enhanced motility (e.g., fliC and pilD genes) and energy metabolism (e.g., pgm and pgk genes) of carrier bacteria and suppressed the defense system (e.g., MSH3 and FLS2 genes) and energy metabolism (e.g., petB and petC genes) of pathogens. Metabolomics analysis revealed that the phoresy system stimulated the secretion of beneficial metabolites (e.g., flavonoid and tropane alkaloid) that could enhance stress response and phyllosphere protection in soybeans. Overall, the phoresy of phages hitchhiking on biocontrol bacteria offers a novel and effective strategy for phyllosphere microbiome manipulation and bacterial disease control.

|
Scooped by
?
May 4, 5:26 PM
|
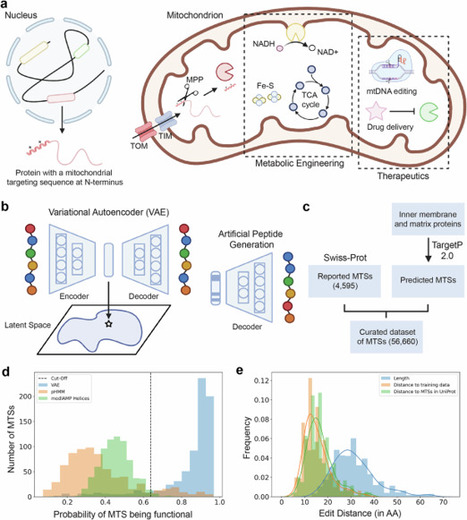
Mitochondria play a key role in energy production and metabolism, making them a promising target for metabolic engineering and disease treatment. However, despite the known influence of passenger proteins on localization efficiency, only a few protein-localization tags have been characterized for mitochondrial targeting. To address this limitation, we leverage a Variational Autoencoder to design novel mitochondrial targeting sequences. In silico analysis reveals that a high fraction of the generated peptides (90.14%) are functional and possess features important for mitochondrial targeting. We characterize artificial peptides in four eukaryotic organisms and, as a proof-of-concept, demonstrate their utility in increasing 3-hydroxypropionic acid titers through pathway compartmentalization and improving 5-aminolevulinate synthase delivery by 1.62-fold and 4.76-fold, respectively. Moreover, we employ latent space interpolation to shed light on the evolutionary origins of dual-targeting sequences. Overall, our work demonstrates the potential of generative artificial intelligence for both fundamental research and practical applications in mitochondrial biology. Mitochondria play a key role in cellular metabolism. Here, authors develop a Variational Autoencoder to design novel mitochondrial targeting sequences, validating them across several eukaryotic organisms and demonstrating their utility in metabolic engineering and protein delivery.

|
Scooped by
?
May 4, 5:02 PM
|
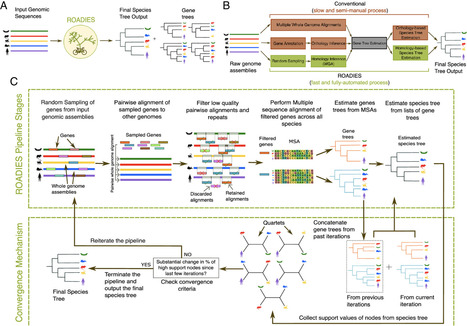
Current genome sequencing initiatives across a wide range of life forms offer significant potential to enhance our understanding of evolutionary relationships and support transformative biological and medical applications. Species trees play a central role in many of these applications; however, despite the widespread availability of genome assemblies, accurate inference of species trees remains challenging due to the limited automation, substantial domain expertise, and computational resources required by conventional methods. To address this limitation, we present ROADIES, a fully automated pipeline to infer species trees starting from raw genome assemblies. In contrast to the prominent approach, ROADIES incorporates a unique strategy of randomly sampling segments of the input genomes to generate gene trees. This eliminates the need for predefining a set of loci, limiting the analyses to a fixed number of genes, and performing the cumbersome gene annotation and/or whole genome alignment steps. ROADIES also eliminates the need to infer orthology by leveraging existing discordance-aware methods that allow multicopy genes. Using the genomic datasets from large-scale sequencing efforts across four diverse life forms (placental mammals, pomace flies, birds, and budding yeasts), we show that ROADIES infers species trees that are comparable in quality to the state-of-the-art studies but in a fraction of the time and effort, including on challenging datasets with rampant gene tree discordance and complex polyploidy. With its speed, accuracy, and automation, ROADIES has the potential to vastly simplify species tree inference, making it accessible to a broader range of scientists and applications.

|
Scooped by
?
May 4, 4:58 PM
|
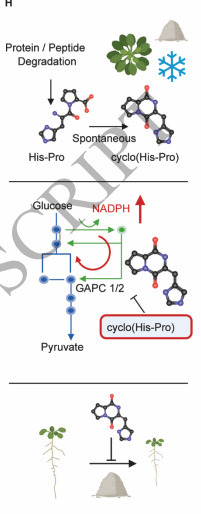
Diketopiperazines (DKPs) are chemically and functionally diverse cyclic dipeptides associated primarily with microbes. Few DKPs have been reported from plants and animals; the best characterized is cyclo(His-Pro), found in the mammalian central nervous system, where it arises from the proteolytic cleavage of a thyrotropin-releasing tripeptide hormone. Herein, we report the identification of cyclo(His-Pro) in Arabidopsis (Arabidopsis thaliana), where its levels increase upon abiotic stress conditions, including high salt, heat, and cold. To screen for potential protein targets, we used isothermal shift assays (iTSA), which examine changes in protein melting stability upon ligand binding. Among the identified proteins, we focused on the glycolytic enzyme, cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC1). Binding between the GAPC1 protein and cyclo(His-Pro) was validated using nano differential scanning fluorimetry (nanoDSF) and microscale thermophoresis (MST), and we could further demonstrate that cyclo(His-Pro) inhibits GAPC1 activity with an IC50 of approximately 200 μM. This inhibition was conserved in human GAPDH. Inhibition of glyceraldehyde-3-phosphate dehydrogenase activity has previously been reported to reroute carbon from glycolysis towards the pentose phosphate pathway. Accordingly, cyclo(His-Pro) supplementation augmented NADPH levels, increasing the NADPH/NADP+ ratio. Phenotypic screening revealed that plants supplemented with cyclo(His-Pro) were more tolerant to high salt stress, as manifested by higher biomass, which we show is dependent on GAPC1/2. Our work reports the identification and functional characterization of cyclo(His-Pro) as a modulator of glyceraldehyde-3-phosphate dehydrogenase in plants.
|



 Your new post is loading...
Your new post is loading...